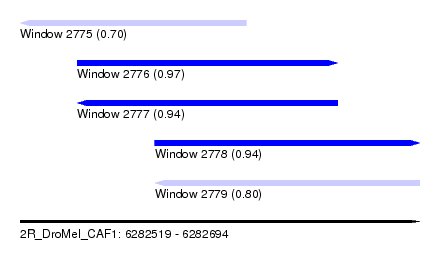
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,282,519 – 6,282,694 |
| Length | 175 |
| Max. P | 0.966536 |

| Location | 6,282,519 – 6,282,618 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.13 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -11.22 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6282519 99 - 20766785 GUCACCCACACUAUCCAUGAACUUAUUUAUCUAUGGUAUAGAUAUAUGGGCAUCUGGAGGUGACAGACAUAUU-UUUAAGGCUACUU---------------AAUUCCACACUCG ((((((.......((((.((.(((((.(((((((...))))))).)))))..)))))))))))).........-.(((((....)))---------------))........... ( -23.01) >DroSec_CAF1 25111 95 - 1 GUCACCCACACUAUCCAUAAACUUAUUUAUCUAUGAGAUAGAUAUUUUGGUAUCUGCAGCUGACUGACGGAUU-UUUAAGGCUAU-------------------CGCCACAUUUA ..............(((......((((((((.....))))))))...))).((((((((....))).))))).-.....(((...-------------------.)))....... ( -17.90) >DroEre_CAF1 24891 106 - 1 GUCACCCACACUUUCCUGCAACCUAUAGA---------UAUGUAUAUGGGCACCAGGAGGUGACUGAUGGAUUUUUUGAGGCAACCUCCAACAUUUACUUACAAUUCCACACUCG ((((((.......(((((...((((((..---------......))))))...)))))))))))((.((((..((.((((.................)))).)).)))))).... ( -25.14) >consensus GUCACCCACACUAUCCAUAAACUUAUUUAUCUAUG__AUAGAUAUAUGGGCAUCUGGAGGUGACUGACGGAUU_UUUAAGGCUAC_U_______________AAUUCCACACUCG ((((((..((....(((......((((((((.....))))))))...)))....))..))))))................................................... (-11.22 = -12.17 + 0.95)

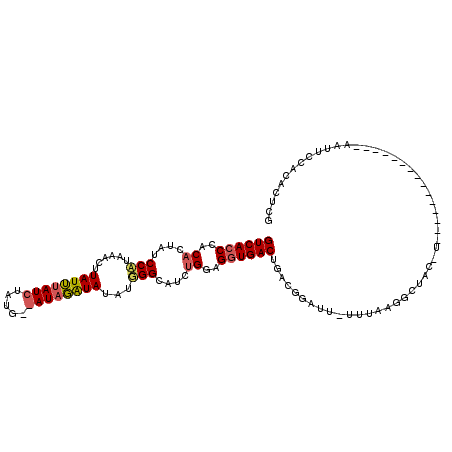
| Location | 6,282,544 – 6,282,658 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -17.19 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6282544 114 + 20766785 A-AAUAUGUCUGUCACCU-----CCAGAUGCCCAUAUAUCUAUACCAUAGAUAAAUAAGUUCAUGGAUAGUGUGGGUGACAAGUCAGCGUAGCAAACUCUGAUUGAUGUUGAUGCAAUUG .-....(((.(((((((.-----.((((((......)))))...((((..((......))..)))).....)..))))))).((((((((..((.....))....))))))))))).... ( -28.00) >DroSec_CAF1 25132 114 + 1 A-AAUCCGUCAGUCAGCU-----GCAGAUACCAAAAUAUCUAUCUCAUAGAUAAAUAAGUUUAUGGAUAGUGUGGGUGACAAGUCAGAGCAGCAAACUCUGAUUGAUGUUGAUGCAAUUG .-.(((((((((((((((-----((....(((...(((.(((((.(((((((......))))))))))))))).)))((....))...)))))......)))))))))..)))....... ( -33.20) >DroEre_CAF1 24931 106 + 1 AAAAUCCAUCAGUCACCU-----CCUGGUGCCCAUAUACAUA---------UCUAUAGGUUGCAGGAAAGUGUGGGUGACAAGUCACAGCAACCAGCUCUGAUUGAUGUUGAUGCAAUUG ......((((((((((((-----((((..(((.(((......---------..))).)))..))))).......)))))).(((((.(((.....))).))))).....)))))...... ( -29.81) >DroYak_CAF1 28383 105 + 1 A-AAUCCAUCAGUCACCUACAGUCCAGGUCC-----UACACA---------UCUAUAAGUACAUGGAAAGUGUGGGUGACAAGUCAAAGCAACAAACUCUGAUUGAUGUUGAUGCAAUUG .-....(((((((((((((((.((((.((.(-----(.....---------......)).)).))))...)))))))))).(((((.((.......)).))))).....)))))...... ( -29.20) >consensus A_AAUCCAUCAGUCACCU_____CCAGAUGCCCAUAUACAUA_________UAAAUAAGUUCAUGGAAAGUGUGGGUGACAAGUCAGAGCAACAAACUCUGAUUGAUGUUGAUGCAAUUG ......((((.(((((((.....(((...((...........................))...))).......))))))).(((((((((.....)))))))))))))............ (-17.19 = -18.13 + 0.94)

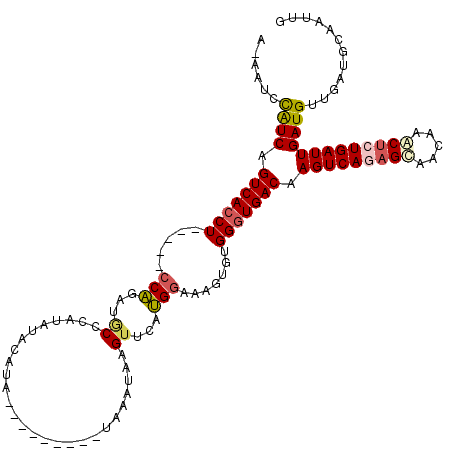
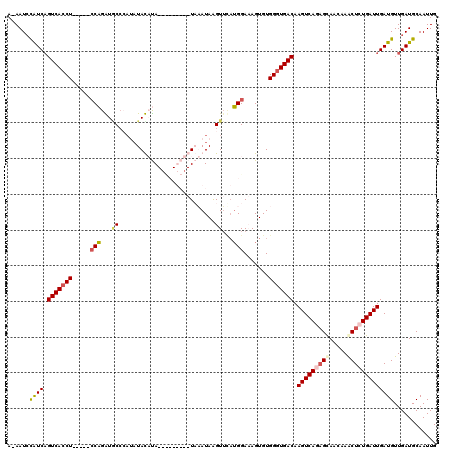
| Location | 6,282,544 – 6,282,658 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.96 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6282544 114 - 20766785 CAAUUGCAUCAACAUCAAUCAGAGUUUGCUACGCUGACUUGUCACCCACACUAUCCAUGAACUUAUUUAUCUAUGGUAUAGAUAUAUGGGCAUCUGG-----AGGUGACAGACAUAUU-U ..................((((.((.....)).)))).((((((((.......((((.((.(((((.(((((((...))))))).)))))..)))))-----))))))))).......-. ( -26.51) >DroSec_CAF1 25132 114 - 1 CAAUUGCAUCAACAUCAAUCAGAGUUUGCUGCUCUGACUUGUCACCCACACUAUCCAUAAACUUAUUUAUCUAUGAGAUAGAUAUUUUGGUAUCUGC-----AGCUGACUGACGGAUU-U .......(((((...((((((((((.....))))))).))).........(((((((((............)))).))))).....)))))((((((-----((....))).))))).-. ( -24.20) >DroEre_CAF1 24931 106 - 1 CAAUUGCAUCAACAUCAAUCAGAGCUGGUUGCUGUGACUUGUCACCCACACUUUCCUGCAACCUAUAGA---------UAUGUAUAUGGGCACCAGG-----AGGUGACUGAUGGAUUUU ............(((((.(((.(((.....))).)))...((((((.......(((((...((((((..---------......))))))...))))-----))))))))))))...... ( -32.91) >DroYak_CAF1 28383 105 - 1 CAAUUGCAUCAACAUCAAUCAGAGUUUGUUGCUUUGACUUGUCACCCACACUUUCCAUGUACUUAUAGA---------UGUGUA-----GGACCUGGACUGUAGGUGACUGAUGGAUU-U ............(((((.(((((((.....)))))))...((((((.(((...((((.((.((((((..---------..))))-----)))).)))).))).)))))))))))....-. ( -32.80) >consensus CAAUUGCAUCAACAUCAAUCAGAGUUUGCUGCUCUGACUUGUCACCCACACUAUCCAUGAACUUAUAGA_________UAGAUAUAUGGGCACCUGG_____AGGUGACUGACGGAUU_U ............((((..(((((((.....)))))))...((((((........(((....((((((((((.....))))))))....))....)))......)))))).))))...... (-15.72 = -16.96 + 1.25)

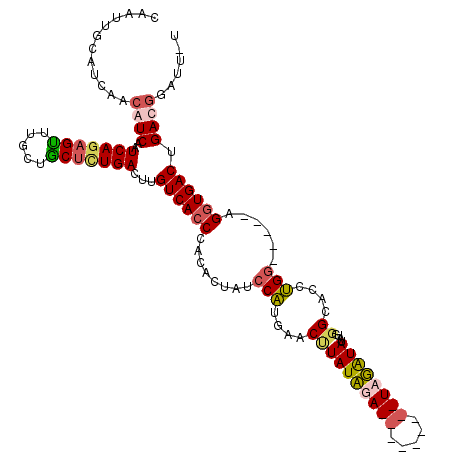
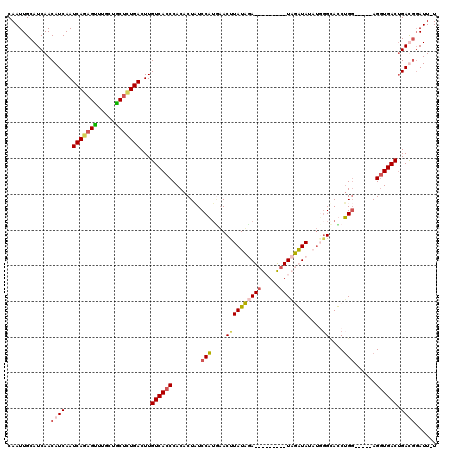
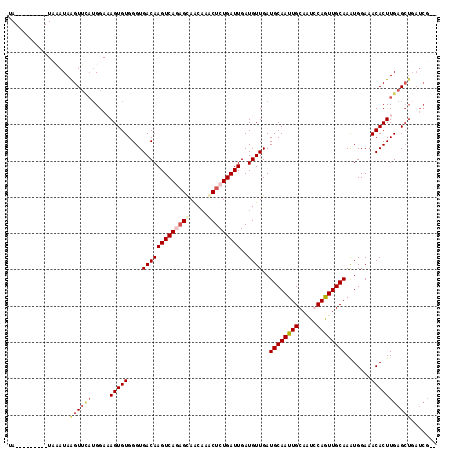
| Location | 6,282,578 – 6,282,694 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -21.31 |
| Energy contribution | -23.25 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6282578 116 + 20766785 UAUACCAUAGAUAAAUAAGUUCAUGGAUAGUGUGGGUGACAAGUCAGCGUAGCAAACUCUGAUUGAUGUUGAUGCAAUUGCAAUGCAAUUGCAAAUGGAAACACUUGAGCUGAUCG-- ....((((..((......))..)))).((((.((((((....((((((((..((.....))....))))))))((((((((...)))))))).........)))))).))))....-- ( -30.40) >DroSec_CAF1 25166 109 + 1 UAUCUCAUAGAUAAAUAAGUUUAUGGAUAGUGUGGGUGACAAGUCAGAGCAGCAAACUCUGAUUGAUGUUGAUGCAAUUGCAAUGCAGUUGCAAAUGGAAACACUUGAG--------- ((((.(((((((......)))))))))))((((....((((((((((((.......))))))))..))))..(((((((((...))))))))).......)))).....--------- ( -32.70) >DroEre_CAF1 24966 109 + 1 UA---------UCUAUAGGUUGCAGGAAAGUGUGGGUGACAAGUCACAGCAACCAGCUCUGAUUGAUGUUGAUGCAAUUGCAAUCCAGUUGCAAGUGGAAACACUUGAGCUGAUCGCC ..---------......((((((((....(((..(....)....))).(((..((((((.....)).)))).)))..))))))))(((((.((((((....)))))))))))...... ( -37.00) >DroYak_CAF1 28417 107 + 1 CA---------UCUAUAAGUACAUGGAAAGUGUGGGUGACAAGUCAAAGCAACAAACUCUGAUUGAUGUUGAUGCAAUUGCAGUCCAGUUGCAAAUGGAAACACUUGAGCUGAUCG-- ((---------((((((..(.......)..))))))))....(((((..(((((.....)).)))...)))))((....)).(..(((((.(((.((....)).))))))))..).-- ( -22.90) >consensus UA_________UAAAUAAGUUCAUGGAAAGUGUGGGUGACAAGUCAGAGCAACAAACUCUGAUUGAUGUUGAUGCAAUUGCAAUCCAGUUGCAAAUGGAAACACUUGAGCUGAUCG__ .................((((((.....(((((....(((((((((((((.....)))))))))..))))..((((((((.....)))))))).......)))))))))))....... (-21.31 = -23.25 + 1.94)

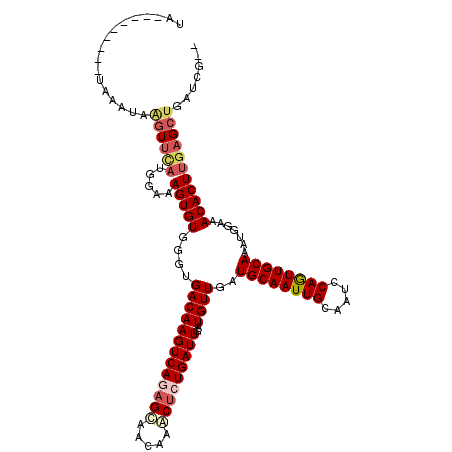
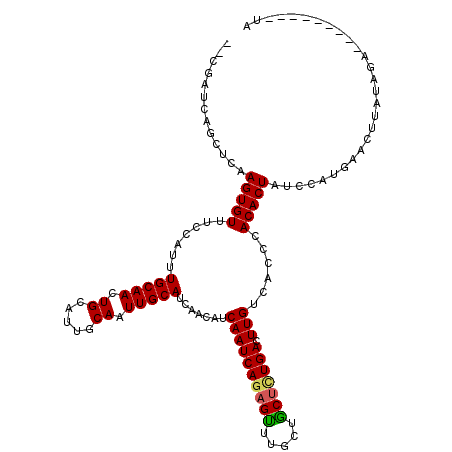
| Location | 6,282,578 – 6,282,694 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -16.95 |
| Energy contribution | -16.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6282578 116 - 20766785 --CGAUCAGCUCAAGUGUUUCCAUUUGCAAUUGCAUUGCAAUUGCAUCAACAUCAAUCAGAGUUUGCUACGCUGACUUGUCACCCACACUAUCCAUGAACUUAUUUAUCUAUGGUAUA --.(((.((.(((.((((...((..(((((((((...)))))))))..(((.((.....)))))))..))))))))).)))......(((((..(((((....)))))..)))))... ( -22.40) >DroSec_CAF1 25166 109 - 1 ---------CUCAAGUGUUUCCAUUUGCAACUGCAUUGCAAUUGCAUCAACAUCAAUCAGAGUUUGCUGCUCUGACUUGUCACCCACACUAUCCAUAAACUUAUUUAUCUAUGAGAUA ---------....(((((.......(((((.(((...))).))))).......((((((((((.....))))))).)))......))))).........(((((......)))))... ( -23.60) >DroEre_CAF1 24966 109 - 1 GGCGAUCAGCUCAAGUGUUUCCACUUGCAACUGGAUUGCAAUUGCAUCAACAUCAAUCAGAGCUGGUUGCUGUGACUUGUCACCCACACUUUCCUGCAACCUAUAGA---------UA (((((((((((((((((....)))))((((.((.....)).))))..............))))))))))))((((....))))........................---------.. ( -31.60) >DroYak_CAF1 28417 107 - 1 --CGAUCAGCUCAAGUGUUUCCAUUUGCAACUGGACUGCAAUUGCAUCAACAUCAAUCAGAGUUUGUUGCUUUGACUUGUCACCCACACUUUCCAUGUACUUAUAGA---------UG --..(((....((((((....))))))..((((((.(((....))).......((((((((((.....))))))).)))............)))).)).......))---------). ( -17.90) >consensus __CGAUCAGCUCAAGUGUUUCCAUUUGCAACUGCAUUGCAAUUGCAUCAACAUCAAUCAGAGUUUGCUGCUCUGACUUGUCACCCACACUAUCCAUGAACUUAUAGA_________UA .............(((((.......(((((.((.....)).))))).......((((((((((.....))))))).)))......)))))............................ (-16.95 = -16.95 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:06 2006