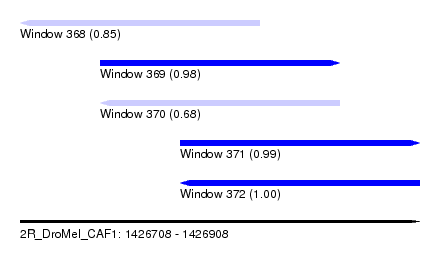
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,426,708 – 1,426,908 |
| Length | 200 |
| Max. P | 0.999070 |

| Location | 1,426,708 – 1,426,828 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -36.28 |
| Energy contribution | -36.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1426708 120 - 20766785 AUGUAAAUGAAAUGCCACUCGGCAGAGUGGCUUACCCUUUGGAAGAAGCUUCCUCUGAGAAGUUAACUGAGGUGAGGUGCCAGAACAGAUUGGUCGGACGGCCAUUGUCUGGGUCGACAA .(((.........(((((((....)))))))..(((((((((.....(((((.(((.((.......)).))).))))).)))))..(((((((((....)))))..))))))))..))). ( -40.80) >DroSec_CAF1 45149 120 - 1 AUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGAAGGAGCUUCCUCCGAGAAGUUAACUGAGGUGAGGUGCCAGAACAGAUUGGUCGGACGGCCAUUGUCUGGGGCGACAG .(((.........(((((((....)))))))(((((.((((((.((.....)))))))).((....))..))))).((.(((((.(((..(((((....))))))))))))).)).))). ( -44.00) >DroSim_CAF1 50205 120 - 1 AUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGAAGGAGCUUCCUCCGAGAAGUUAACUGAGGUGAGGUGCCAGAACAGAUUGGUCGGACGGCCAUUGUCUGGGUCGACAA .(((..((.....(((((((....)))))))(((((.((((((.((.....)))))))).((....))..)))))....(((((.(((..(((((....)))))))))))))))..))). ( -40.40) >DroEre_CAF1 43252 120 - 1 AUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGGAGGAGCUGCCUCCGAGAAGUUAAGCGAGGUGAGGUGCCAGAACAGAUUGGUCGGACGUCCAUUGUCUGGGUCGACAA .(((..((.....(((((((....)))))))(((((((((.(((((.....))))).))).(.....)).)))))....(((((.(((..(((.(....).)))))))))))))..))). ( -39.30) >DroYak_CAF1 47811 120 - 1 AUGUAAAUGAAAUGCCAAUCUGCAGAGUGGCUUACCCUUUGGAAGGAGCUUCUUUCGAGAAGUUAACUGAGGUGAGGUGCCAGAACGGAUUGGUCGGACGGCCAUUGUCUGGGCCGACAA .(((.........(((((((((....(((.((((((..(..(....(((((((....)))))))..)..))))))).))).....)))))))))....(((((........)))))))). ( -39.60) >consensus AUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGAAGGAGCUUCCUCCGAGAAGUUAACUGAGGUGAGGUGCCAGAACAGAUUGGUCGGACGGCCAUUGUCUGGGUCGACAA .(((.........(((((((....))))))).((((((((((....((((((......))))))..))))))...))))(((((.(((..(((((....)))))))))))))....))). (-36.28 = -36.52 + 0.24)

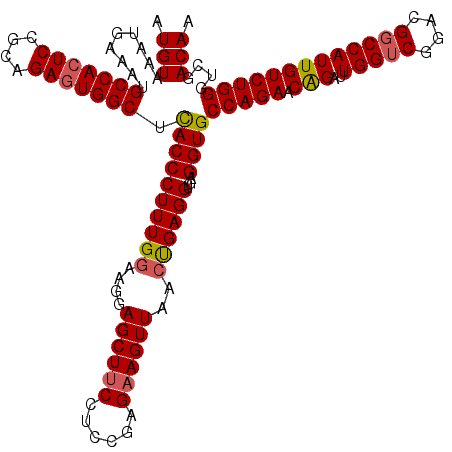
| Location | 1,426,748 – 1,426,868 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1426748 120 + 20766785 GCACCUCACCUCAGUUAACUUCUCAGAGGAAGCUUCUUCCAAAGGGUAAGCCACUCUGCCGAGUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUAUUUU ...............((((...((((.(((((...)))))...((((..(((((((....))))))).........((((....)))).))))((.....)).)))).....)))).... ( -30.10) >DroSec_CAF1 45189 120 + 1 GCACCUCACCUCAGUUAACUUCUCGGAGGAAGCUCCUUCCAAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCGCCCGCUUUCGGCACUGACAGUCGUUACUUU ..........(((((.........((((....)))).......(((((((((((((....)))))))....((((........))))))))))((.....)))))))............. ( -35.60) >DroSim_CAF1 50245 120 + 1 GCACCUCACCUCAGUUAACUUCUCGGAGGAAGCUCCUUCCAAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCGCCCGCUUUCGGCACUGACAGUCGUUACUUU ..........(((((.........((((....)))).......(((((((((((((....)))))))....((((........))))))))))((.....)))))))............. ( -35.60) >DroEre_CAF1 43292 120 + 1 GCACCUCACCUCGCUUAACUUCUCGGAGGCAGCUCCUCCCAAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUAUUUU ............(((.........(((((.....)))))....(((((((((((((....)))))))....((((........))))))))))......)))..((((....)))).... ( -34.50) >DroYak_CAF1 47851 120 + 1 GCACCUCACCUCAGUUAACUUCUCGAAAGAAGCUCCUUCCAAAGGGUAAGCCACUCUGCAGAUUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUGUUUU ..........(((((...(((((....)))))...........((((..((((.((....)).)))).........((((....)))).))))((.....)))))))............. ( -22.40) >consensus GCACCUCACCUCAGUUAACUUCUCGGAGGAAGCUCCUUCCAAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUAUUUU ..........(((((.........(((((.....)))))....(((((((((((((....)))))))....((((........))))))))))((.....)))))))............. (-28.90 = -29.22 + 0.32)

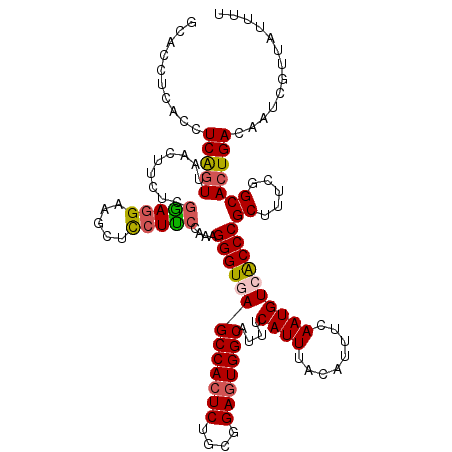
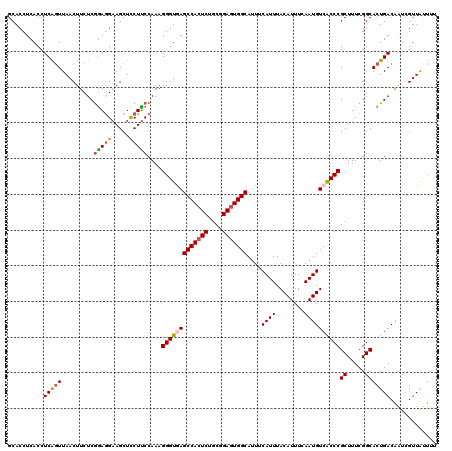
| Location | 1,426,748 – 1,426,868 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1426748 120 - 20766785 AAAAUAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCACUCGGCAGAGUGGCUUACCCUUUGGAAGAAGCUUCCUCUGAGAAGUUAACUGAGGUGAGGUGC .......(...(.(((((.(((((...((((((((((........))))....(((((((....))))))))))))))))))....((((((......)))))).))))).)...).... ( -36.50) >DroSec_CAF1 45189 120 - 1 AAAGUAACGACUGUCAGUGCCGAAAGCGGGCGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGAAGGAGCUUCCUCCGAGAAGUUAACUGAGGUGAGGUGC .......(((.((((.((.((....).).)))))))))...............(((((((....)))))))(((((.((((((.((.....)))))))).((....))..)))))..... ( -34.80) >DroSim_CAF1 50245 120 - 1 AAAGUAACGACUGUCAGUGCCGAAAGCGGGCGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGAAGGAGCUUCCUCCGAGAAGUUAACUGAGGUGAGGUGC .......(((.((((.((.((....).).)))))))))...............(((((((....)))))))(((((.((((((.((.....)))))))).((....))..)))))..... ( -34.80) >DroEre_CAF1 43292 120 - 1 AAAAUAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGGAGGAGCUGCCUCCGAGAAGUUAAGCGAGGUGAGGUGC ................(..((..(.((((((((((((........))))....(((((((....)))))))))))))(((.(((((.....))))).)))......))....)..))..) ( -41.30) >DroYak_CAF1 47851 120 - 1 AAAACAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCAAUCUGCAGAGUGGCUUACCCUUUGGAAGGAGCUUCUUUCGAGAAGUUAACUGAGGUGAGGUGC ....((.(...(.(((((.(((((...((((((((((........))))....((((.((....)).)))))))))))))))....(((((((....))))))).))))).)...).)). ( -31.60) >consensus AAAAUAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUUGGAAGGAGCUUCCUCCGAGAAGUUAACUGAGGUGAGGUGC ................(..((....((((((((((((........))))....(((((((....)))))))))))))(((((.(((.....))))))))............))..))..) (-31.80 = -31.88 + 0.08)

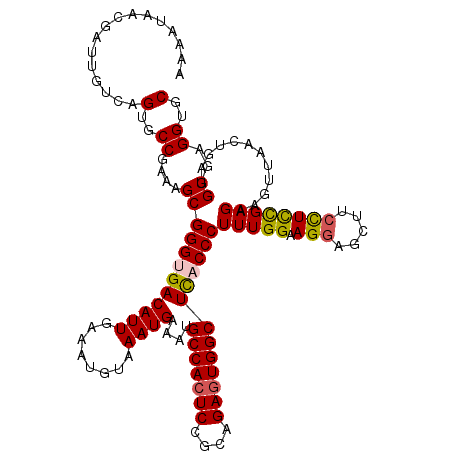
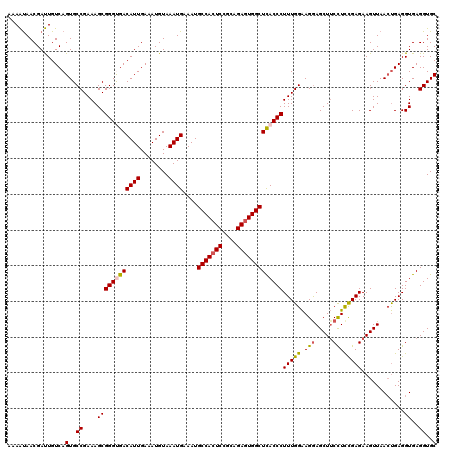
| Location | 1,426,788 – 1,426,908 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -32.04 |
| Energy contribution | -32.44 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1426788 120 + 20766785 AAAGGGUAAGCCACUCUGCCGAGUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUAUUUUCCGUUACAAAUCAAUUGCCAACUUAUGGCCGUGACGAGCC ...((((..(((((((....))))))).........((((....)))).))))......(((..((((....)))).....((((((.........((((.....)))).)))))).))) ( -33.10) >DroSec_CAF1 45229 120 + 1 AAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCGCCCGCUUUCGGCACUGACAGUCGUUACUUUCCGUUACAAAUCAAUUGCCAACUUAUGGCCGUGACGAGCC ...(((((((((((((....)))))))....((((........))))))))))......(((..((((....)))).....((((((.........((((.....)))).)))))).))) ( -36.90) >DroSim_CAF1 50285 120 + 1 AAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCGCCCGCUUUCGGCACUGACAGUCGUUACUUUCCGUUACAAAUCAAUUGCCAACUUAUGGCCGUGACGAGCC ...(((((((((((((....)))))))....((((........))))))))))......(((..((((....)))).....((((((.........((((.....)))).)))))).))) ( -36.90) >DroEre_CAF1 43332 120 + 1 AAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUAUUUUCCGCUACAAAUCAAUUGCCAACUUAUGGCCGUGACGAGCC ...(((((((((((((....)))))))....((((........))))))))))((((.(((...((((....))))....))).......(((...((((.....))))..))).)))). ( -36.80) >DroYak_CAF1 47891 120 + 1 AAAGGGUAAGCCACUCUGCAGAUUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUGUUUUCCGUUGCAAAUCAAUUGCCAUCUUAUGACCGUGACGAGCC ...((((..((((.((....)).)))).........((((....)))).))))((((((((((.(((......((((........)))).)))..)))).((....))....)).)))). ( -24.60) >consensus AAAGGGUGAGCCACUCUGCGGAGUGGCAUUUCAUUUACAUUUCAAUGUCACCCGCUUUCGGCACUGACAAUCGUUAUUUUCCGUUACAAAUCAAUUGCCAACUUAUGGCCGUGACGAGCC ...(((((((((((((....)))))))....((((........))))))))))((((.(((...((((....))))....))).......(((...((((.....))))..))).)))). (-32.04 = -32.44 + 0.40)

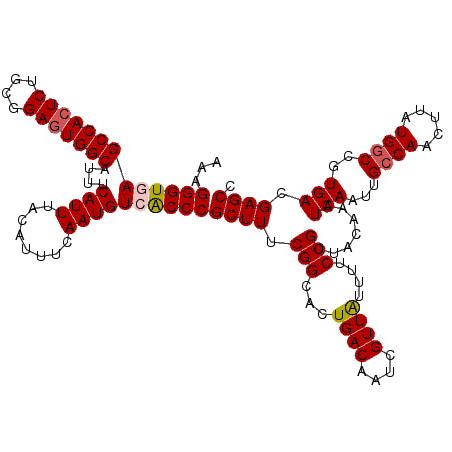
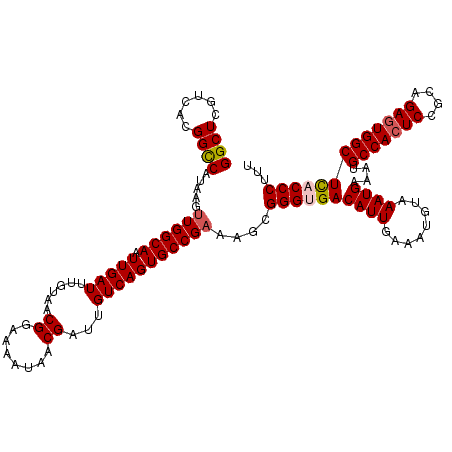
| Location | 1,426,788 – 1,426,908 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -36.24 |
| Energy contribution | -36.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1426788 120 - 20766785 GGCUCGUCACGGCCAUAAGUUGGCAAUUGAUUUGUAACGGAAAAUAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCACUCGGCAGAGUGGCUUACCCUUU ((((......)))).....((((((.(((((......((........))...)))))))))))....((((((((((........))))....(((((((....)))))))))))))... ( -39.50) >DroSec_CAF1 45229 120 - 1 GGCUCGUCACGGCCAUAAGUUGGCAAUUGAUUUGUAACGGAAAGUAACGACUGUCAGUGCCGAAAGCGGGCGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUU ((((......)))).....((((((.(((((((((.((.....)).))))..)))))))))))....(((.((((((........))))....(((((((....))))))))).)))... ( -39.50) >DroSim_CAF1 50285 120 - 1 GGCUCGUCACGGCCAUAAGUUGGCAAUUGAUUUGUAACGGAAAGUAACGACUGUCAGUGCCGAAAGCGGGCGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUU ((((......)))).....((((((.(((((((((.((.....)).))))..)))))))))))....(((.((((((........))))....(((((((....))))))))).)))... ( -39.50) >DroEre_CAF1 43332 120 - 1 GGCUCGUCACGGCCAUAAGUUGGCAAUUGAUUUGUAGCGGAAAAUAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUU ((((......)))).....((((((.(((((......((........))...)))))))))))....((((((((((........))))....(((((((....)))))))))))))... ( -41.60) >DroYak_CAF1 47891 120 - 1 GGCUCGUCACGGUCAUAAGAUGGCAAUUGAUUUGCAACGGAAAACAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCAAUCUGCAGAGUGGCUUACCCUUU ((...(((((.......((((((((.((.(((((((.((........))..(((((.(..(....)..).)))))......))))))).)).)))).)))).....)))))...)).... ( -33.10) >consensus GGCUCGUCACGGCCAUAAGUUGGCAAUUGAUUUGUAACGGAAAAUAACGAUUGUCAGUGCCGAAAGCGGGUGACAUUGAAAUGUAAAUGAAAUGCCACUCCGCAGAGUGGCUCACCCUUU ((((......)))).....((((((.(((((......((........))...)))))))))))....((((((((((........))))....(((((((....)))))))))))))... (-36.24 = -36.64 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:49 2006