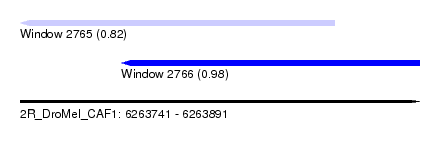
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,263,741 – 6,263,891 |
| Length | 150 |
| Max. P | 0.980784 |

| Location | 6,263,741 – 6,263,859 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.41 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -26.10 |
| Energy contribution | -28.14 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6263741 118 - 20766785 UAUCUGGUUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGCCUGUGAUGCCAUAAAGGCAUUGGUUUAUCAGCUGAAUUCCAGCAUUCAGAA ..(((((..(((...........((((.....)))).(((((((.((((.....))))))))))))))...((((((.....)))))).........((((.....))))..))))). ( -38.90) >DroSec_CAF1 6910 118 - 1 UAUCUGGCUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGUCUGUGAUGCCAUAAAGGCAUUGGCUUAUCAGCUGAAUUCCAGCAUUCGGAA ..(((((.((((((..((..((.((((.....)))).(((((((.((((.....))))))))))))).))..))))))...((((....))))....((((.....))))..))))). ( -39.30) >DroSim_CAF1 7133 118 - 1 UAUCUGGCUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGUCUGUGAUGCCAUAAAGGCAUUGGUUUAUCAGCUGAAUUCCAGCAUUCGGAA ..(((((((...((.((((((((((((.....)))).(((((((.((((.....)))))))))))...)))))))).))...)))............((((.....))))...)))). ( -37.60) >DroEre_CAF1 6782 93 - 1 UAUCCGGCUGGCAUUGUACUGCAGGCUCUUUGGGCCAACAAGAUGCCGAAGCG----------CUGCCUCCGA---------------GGUUUAUCAGCUGAAUUCCAGCAUUCGGCA .((((((..((((..(..((...(((..((((......))))..)))..))..----------))))).))).---------------)))......(((((((......))))))). ( -29.30) >DroYak_CAF1 7729 117 - 1 UAUCUGGCUGGCAUUGUGUUACAGGCUCUUUGGG-CAACAAGAUGCCGAAACUCUUGGCGUCUCUGCCUGUGAUAGCAUAAAGGCAUUGGUUUAUCAGCUGAAUUCCAGCAUUCAGAA ..(((((((.....((((((((((((......(.-...).(((((((((.....)))))))))..)))))))))).))....)))............((((.....))))...)))). ( -41.50) >consensus UAUCUGGCUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGCCUGUGAUGCCAUAAAGGCAUUGGUUUAUCAGCUGAAUUCCAGCAUUCGGAA ..(((((((((.((((((((((((((.....(((((..(((((.........))))))))))...)))))))))))............(((......))).))).)))))...)))). (-26.10 = -28.14 + 2.04)

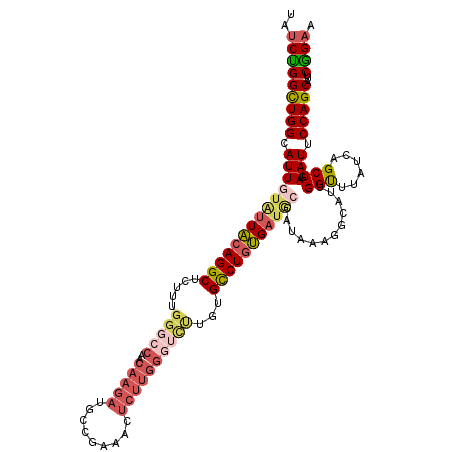
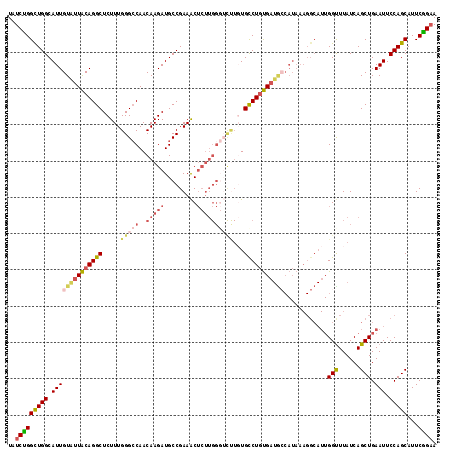
| Location | 6,263,779 – 6,263,891 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.16 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6263779 112 - 20766785 U--------GGGUAUUGUAGUUGGGCUGAAUGGUUUGCGGUAUCUGGUUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGCCUGUGAUGCCAUA .--------.(((((..(((..(((((.((.(((((.((((((((.((((((.(((.....))).........)))))).))))))))))))).)).))))).....)))..)))))... ( -44.60) >DroSec_CAF1 6948 112 - 1 U--------GGGUAUUGUACUUGGGCGGAAUGGUUUGUGGUAUCUGGCUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGUCUGUGAUGCCAUA .--------.(((((..((..(((.(.(((.((((((((((((...(....)...)))))))))))).))).).)))(((((((.((((.....)))))))))))...))..)))))... ( -39.10) >DroSim_CAF1 7171 112 - 1 U--------GGGCAUUGUAGUUGGGCGGAAUGGUUUGUGGUAUCUGGCUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGUCUGUGAUGCCAUA .--------.(((((..(((.(((.(.(((.((((((((((((...(....)...)))))))))))).))).).)))(((((((.((((.....)))))))))))..)))..)))))... ( -43.80) >DroEre_CAF1 6812 94 - 1 ---------GGGCAUUGUAGUUGAGCUGAAUGGUUUGUGGUAUCCGGCUGGCAUUGUACUGCAGGCUCUUUGGGCCAACAAGAUGCCGAAGCG----------CUGCCUCCGA------- ---------.((((((...((((.(((.((.((((((..((((...(....)...))))..))))))..)).)))))))..))))))((.((.----------..)).))...------- ( -32.10) >DroYak_CAF1 7767 119 - 1 UGGUUAACUGGGUAUUGUAGUUCAGCAGAAUGGUUUGUGGUAUCUGGCUGGCAUUGUGUUACAGGCUCUUUGGG-CAACAAGAUGCCGAAACUCUUGGCGUCUCUGCCUGUGAUAGCAUA ..((((.(..(((((..(((..((......))..)))..)))))..).))))..((((((((((((......(.-...).(((((((((.....)))))))))..)))))))))).)).. ( -43.90) >consensus U________GGGUAUUGUAGUUGGGCGGAAUGGUUUGUGGUAUCUGGCUGGCAUUGUAUUACAGGCUCUUUGGGCCAACAAGAUGCCGAAACUCUUGGGUCUUGUGCCUGUGAUGCCAUA ..........((((((...(((((.(..((.((((((((((((...(....)...)))))))))))).))..).)))))..))))))................................. (-26.84 = -27.16 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:52 2006