| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,224,292 – 6,224,412 |
| Length | 120 |
| Max. P | 0.599971 |

| Location | 6,224,292 – 6,224,412 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -40.65 |
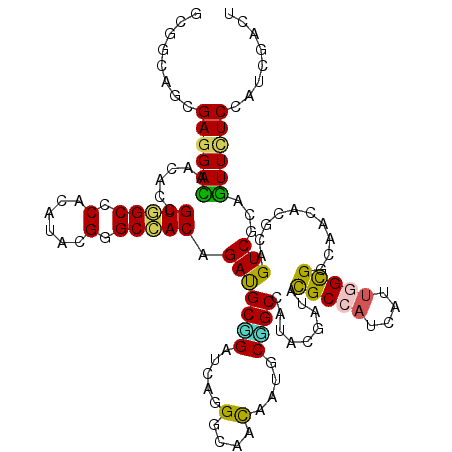
| Consensus MFE | -27.34 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
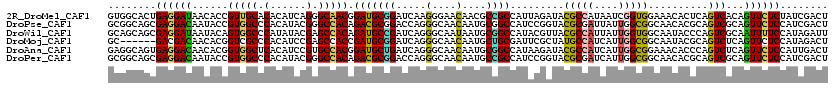
>2R_DroMel_CAF1 6224292 120 + 20766785 GUGGCACUGAGGAUAACACCGUUGCACACAUCAGGGCAACGGAUGCGGAUCAAGGGAACAACGCCGCCAUUAGAUACGCCAUAAUCGGUGGAAACACUCAGUCACAGUUCUCUAUCGACU (((((.............(((((((.(......).)))))))..((((.....(....)....))))..........)))))..(((((((((((...........))).)))))))).. ( -34.40) >DroPse_CAF1 49578 120 + 1 GCGGCAGCGAGGACAAUACCGUGGCCCACAUACGGGCCACAGACGCGGACCAGGGCAACAAUGCGGCCAUCCGGUACGCGAUUAUUGGCGGCAACACGCAGUCGCAGUUCUCCAUCGACU ((....))((((((..(((((((((((......)))))))......(((....(((.........))).))))))).((((((....(((......))))))))).))))))........ ( -47.20) >DroWil_CAF1 51947 120 + 1 GCAGCAGCGAGGAUAAUACAGUGGCCCAUAUACGAGCCACAGAUGCCGAUCAGGGCAAUAAUGCGGCCAUACGUUACGCCAUUAUUGGUGGCAAUACCCAGUCGCAAUUUUCCAUAGAUU ((....))..(((.(((...(((((.(......).)))))...((((......))))....((((((.....(((.(((((....)))))..))).....))))))))).)))....... ( -33.90) >DroMoj_CAF1 93503 114 + 1 GC------GACGACAACACGGUCGCCCACAUCCGAGCCACCGAUGCGGAUCAGGGCAACAAUGCUGCGAUUCGCUAUGCCAUCAUUGGCGGCAAUACGCAGUCUCAGUUCUCCAUAGACU ((------(.((((......))))...........(((......(((((((..((((....))))..)))))))...((((....)))))))....)))(((((...........))))) ( -36.30) >DroAna_CAF1 67352 120 + 1 GAGGCAGUGAGGACAACACGGUGGCUCACAUCCGUGCCACGGAUGCUGAUCAGGGCAACAAUGCGGCCAUAAGAUACGCCAUCAUUGGCGGAAACACCCAGUCUCAGUUCUCCAUUGACU .((.((((((((((.....(((.(((((((((((.....)))))).)))....(....)...)).)))....((.((((((....))))(....).....)).)).))))).))))).)) ( -41.50) >DroPer_CAF1 49275 120 + 1 GCGGCAGCGAGGACAAUACCGUGGCCCACAUACGGGCCACAGACGCGGACCAGGGCAACAAUGCCGCCAUCCGGUACGCGAUCAUUGGCGGCAACACGCAGUCGCAGUUCUCCAUCGACU ((....))((((((......(((((((......))))))).((((((......(....)..((((((((...(((.....)))..))))))))...))).)))...))))))........ ( -50.60) >consensus GCGGCAGCGAGGACAACACCGUGGCCCACAUACGGGCCACAGAUGCGGAUCAGGGCAACAAUGCGGCCAUACGAUACGCCAUCAUUGGCGGCAACACGCAGUCGCAGUUCUCCAUCGACU ........((((((......(((((.(......).))))).(((((((.....(....)....)))).........(((((....)))))..........)))...))))))........ (-27.34 = -26.52 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:42 2006