
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
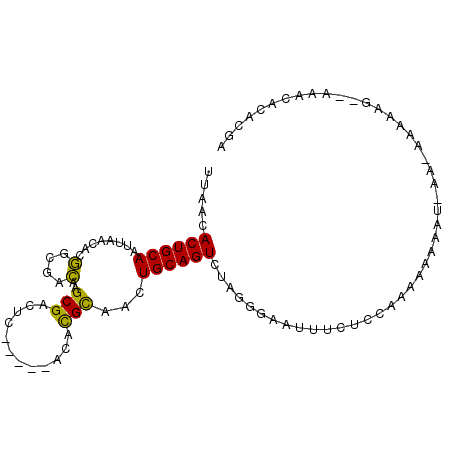
| Location | 6,220,605 – 6,220,699 |
| Length | 94 |
| Max. P | 0.959152 |

| Location | 6,220,605 – 6,220,699 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 70.63 |
| Mean single sequence MFE | -17.58 |
| Consensus MFE | -7.80 |
| Energy contribution | -6.92 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6220605 94 + 20766785 UUAACACUGCAAUUAACGCGGCGACAAGCGUCUC-----ACACGCAACUGCAGUCUGGGGAAUUUCUCCAAAAAUAUA-CAAAAAAAGCAAAACACACGA .....((((((........(....)..((((...-----..))))...)))))).(((((.....)))))........-..................... ( -18.20) >DroPse_CAF1 45793 80 + 1 UUAACACUGCAAUUAACAGAAAGUUAAGCGACUGCCACGACAUGUAAUUGCAGUCUUCAGACUUUCUCCCCGAAAAAC-AC------------------- .................((((((((....((((((....(....)....))))))....))))))))...........-..------------------- ( -15.00) >DroSec_CAF1 51683 92 + 1 UUAACACUGCAAUUAACACGGCGACAAGCGCCUC-----ACACGCAACUGCAGUCUAGGGGAUUUCUCCCAAAAAAAU-AACAAAAAG--AAACACCCGA .....((((((........((((.....))))..-----.........))))))...(((..(((((...........-.......))--)))..))).. ( -19.18) >DroSim_CAF1 53142 92 + 1 UUAACACUGCAAUUAACAUGGCGACAAGCGCCUC-----ACACGCAACUGCAGUCUAGGGGAUUUCUCCAAAAAAAAU-AACAAAAAG--AAACACACGA .....((((((........((((.....))))..-----.........))))))...((((....)))).........-.........--.......... ( -17.21) >DroEre_CAF1 53989 92 + 1 UUAACACUGCAAUUAACACGGCGACAAGCGACUC-----ACACGCAACUGCAGUCUGGGGGAUUUCUCCAAAAA-AAUAACAAAAAUG--GAACCCAGGA .....((((((........(....)..(((....-----...)))...))))))((((((((....))).....-.....((....))--...))))).. ( -20.90) >DroPer_CAF1 45441 80 + 1 UUAACACUGCAAUUAACAGAAAGUUAAGCGACUGCCACGACAUGUAAUUGCAGUCUUCAGACUUUCUCCCCGAAAAAC-AC------------------- .................((((((((....((((((....(....)....))))))....))))))))...........-..------------------- ( -15.00) >consensus UUAACACUGCAAUUAACACGGCGACAAGCGACUC_____ACACGCAACUGCAGUCUAGGGAAUUUCUCCAAAAAAAAU_AA_AAAAAG__AAACACACGA .....((((((........(....)..(((............)))...)))))).............................................. ( -7.80 = -6.92 + -0.89)

| Location | 6,220,605 – 6,220,699 |
|---|---|
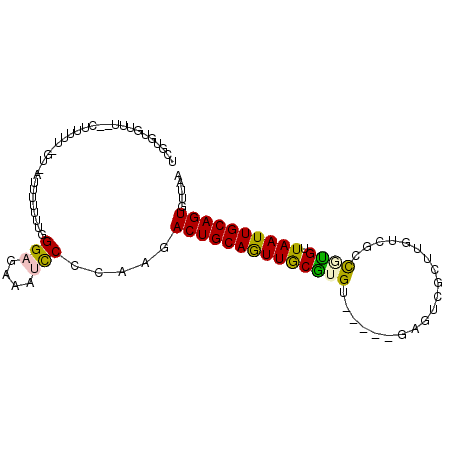
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 70.63 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.17 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6220605 94 - 20766785 UCGUGUGUUUUGCUUUUUUUG-UAUAUUUUUGGAGAAAUUCCCCAGACUGCAGUUGCGUGU-----GAGACGCUUGUCGCCGCGUUAAUUGCAGUGUUAA .....................-.......((((.(.....).))))((((((((((((((.-----(.(((....))).))))).))))))))))..... ( -23.80) >DroPse_CAF1 45793 80 - 1 -------------------GU-GUUUUUCGGGGAGAAAGUCUGAAGACUGCAAUUACAUGUCGUGGCAGUCGCUUAACUUUCUGUUAAUUGCAGUGUUAA -------------------.(-((......((.(((((((.(((.((((((...(((.....)))))))))..)))))))))).))....)))....... ( -19.10) >DroSec_CAF1 51683 92 - 1 UCGGGUGUUU--CUUUUUGUU-AUUUUUUUGGGAGAAAUCCCCUAGACUGCAGUUGCGUGU-----GAGGCGCUUGUCGCCGUGUUAAUUGCAGUGUUAA ..(((..(((--(((((....-........))))))))..)))...((((((((((((...-----..((((.....)))).)).))))))))))..... ( -28.20) >DroSim_CAF1 53142 92 - 1 UCGUGUGUUU--CUUUUUGUU-AUUUUUUUUGGAGAAAUCCCCUAGACUGCAGUUGCGUGU-----GAGGCGCUUGUCGCCAUGUUAAUUGCAGUGUUAA ..........--.........-.........(((....))).....((((((((((((((.-----(.(((....))).))))).))))))))))..... ( -23.10) >DroEre_CAF1 53989 92 - 1 UCCUGGGUUC--CAUUUUUGUUAUU-UUUUUGGAGAAAUCCCCCAGACUGCAGUUGCGUGU-----GAGUCGCUUGUCGCCGUGUUAAUUGCAGUGUUAA ..(((((...--((....)).....-.....(((....))))))))((((((((((((.((-----((........)))))))...)))))))))..... ( -27.50) >DroPer_CAF1 45441 80 - 1 -------------------GU-GUUUUUCGGGGAGAAAGUCUGAAGACUGCAAUUACAUGUCGUGGCAGUCGCUUAACUUUCUGUUAAUUGCAGUGUUAA -------------------.(-((......((.(((((((.(((.((((((...(((.....)))))))))..)))))))))).))....)))....... ( -19.10) >consensus UCGUGUGUUU__CUUUUU_GU_AUUUUUUUGGGAGAAAUCCCCAAGACUGCAGUUGCGUGU_____GAGUCGCUUGUCGCCGUGUUAAUUGCAGUGUUAA ...............................(((....))).....((((((((((((((....................)))).))))))))))..... (-13.86 = -13.17 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:40 2006