| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,218,658 – 6,218,765 |
| Length | 107 |
| Max. P | 0.624996 |

| Location | 6,218,658 – 6,218,765 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.60 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6218658 107 - 20766785 AAUACCGAAAGAAUCGAAAGAAGAAAAGUCUCCAUUGUUGUUGCUGAUGCUGUAGUACUCGUACUUUGCCAAAGGCAGAACAGCAACAAGCAGCUACAAAUGCAAAC .....(....)..((....)).............((((.((((((..((((((.(((....)))((((((...))))))))))))...)))))).))))........ ( -30.40) >DroSec_CAF1 49681 107 - 1 AAUACCGAAAGAAUCGAAAGAAGAAAAGUCUCCAUUGUUGUUGCUGAUGCUGUAGUACUCAUACUUAGCCAAAGGCAGAACAGCAACAAGCAGCUACAAAUGCAAAC .....(....)..((....)).............((((.((((((..((((((((((....))))..(((...)))...))))))...)))))).))))........ ( -27.00) >DroSim_CAF1 51150 107 - 1 AAUACCGAAAGAAUCGAAAGAAGAAAAGUCUCUAUUGUUGUUGCUGAUGCUGUAGUACUCAUACUUAGCCAAAGGCAGAACAGCAACAAGCAGCUACAAAUGCAAAC .....(....)..((....))(((......))).((((.((((((..((((((((((....))))..(((...)))...))))))...)))))).))))........ ( -27.10) >DroEre_CAF1 52060 107 - 1 AAUACCGAGAGAAUCGAAAGAAGAAAAGUCUCCAUUGUUGCUGCUGAUGCUGUAGUACUCAUACUUUGCCAAAGGCUGAACAGCAACAAGCAGCUACAAAUGCAACU ......(.((((.((....)).......))))).((((.((((((..((((((((((....))))..(((...)))...))))))...)))))).))))........ ( -29.90) >DroYak_CAF1 53008 107 - 1 AAUACCAAAAGAAUCGAAAGAAGAAAAGUCUUCAUUGUUGUUGCUGAUGCUGUACUACUCAUACUUCGCCAAAGGCGGAACAGCACCAAGCAGCUACAAAUGCAAAC ...................(((((....))))).((((.((((((..((((((...........((((((...))))))))))))...)))))).))))........ ( -29.30) >DroAna_CAF1 60413 91 - 1 AAUGUUGA-ACAAUCGAAGGAA-----GUUUCCUCUGCUUCUGCUGA------ACU----AUACUUUGCCAACGGCAGAAAAGCAACAAGCAGCUACAAAUGCAAAC ..(((((.-....(((..((((-----((.......))))))..)))------...----....((((((...))))))....))))).(((........))).... ( -20.60) >consensus AAUACCGAAAGAAUCGAAAGAAGAAAAGUCUCCAUUGUUGUUGCUGAUGCUGUAGUACUCAUACUUUGCCAAAGGCAGAACAGCAACAAGCAGCUACAAAUGCAAAC .............((....)).............((((.((((((..((((((((((....))))..(((...)))...))))))...)))))).))))........ (-19.66 = -21.43 + 1.78)

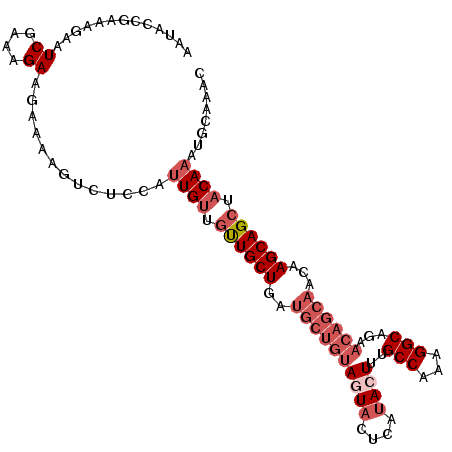

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:39 2006