| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,170,731 – 6,170,835 |
| Length | 104 |
| Max. P | 0.721246 |

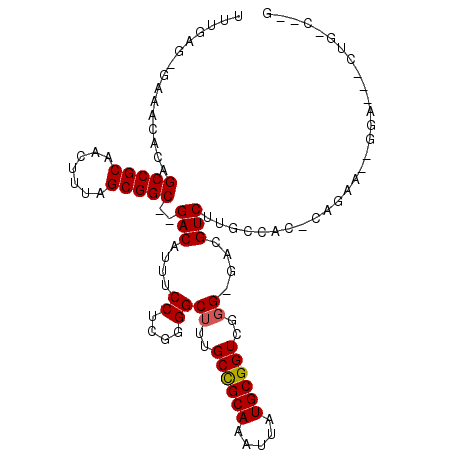
| Location | 6,170,731 – 6,170,835 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6170731 104 + 20766785 UUGGUGGGAAACACAGCCGCAACUUUAGCGGC--GACAUUUCCUCAGGGCUUUGCCGCAAAUUAUGCGGUCGGG-GACGUCUUGCCAC-CAGAA--GGAACUCUGACAGG (((((((((((....(((((.......)))))--....))))).((((((...((((((.....)))))).(..-..))))))).)))-)))..--.............. ( -38.70) >DroPse_CAF1 7722 96 + 1 UUUGAG-GAAACACAGCCGCAACUUUAGCGGCGGGACAUUUCCUCUGGGCUUUGCUGCAAAUUAUGCGGUGGGGCCAUGUCUUGGGGCACGGAA--GGA----------- ...(((-((((..(.(((((.......))))).)....)))))))..(((((..(((((.....)))))..))))).((((....))))(....--)..----------- ( -40.90) >DroEre_CAF1 12003 105 + 1 UUGGUG-GAAACACAGCCGCAACUUUAGCGGC--GACAUUUCCUCGGGGCUUUGCCGCAAAUUAUGCGGUCGGG-GGCGUCUUGCCUC-CAGACUGGGAACACCGCCUUG ..((((-(.......(((((.......)))))--.....((((.(((......((((((.....)))))).(((-(((.....)))))-)...)))))))..)))))... ( -45.00) >DroYak_CAF1 8983 101 + 1 UUCGAG-GAAACACAGCCGCAACUUUAGCGGC--GACAUUUCCUCGGGGCUUUGCCGCAAAUUAUGCGGUCGGG-GACGUCUUGCCAC-CAGAA--GGA--ACUGCCUGG .(((((-((((....(((((.......)))))--....)))))))))(((...((((((.....))))))((..-..))....))).(-(((..--...--.....)))) ( -41.70) >DroAna_CAF1 12180 103 + 1 UUUGAG-GCAACACAGCCGCAACUUUAGCGGC--GACAUUUCCUCCGGGCAUUGCCGCAAAUUAUGCGGUCGGG-GAUGUCUUGCCAC-CAAAA--GGAGCUGUCUCCAG ((((.(-((((....(((((.......)))))--(((((..((.((.(((...)))(((.....)))))..)).-.))))))))))..-)))).--((((....)))).. ( -41.80) >DroPer_CAF1 7744 96 + 1 UUUGAG-GAAACACAGCCGCAACUUUAGCGGCGGGACAUUUCCUCUGGGCUUUGCUGCAAAUUAUGCGGUGGGGCCAUGUCUUGGGGUACGGAA--GGA----------- ...(((-((((..(.(((((.......))))).)....)))))))..(((((..(((((.....)))))..))))).............(....--)..----------- ( -38.30) >consensus UUUGAG_GAAACACAGCCGCAACUUUAGCGGC__GACAUUUCCUCGGGGCUUUGCCGCAAAUUAUGCGGUCGGG_GACGUCUUGCCAC_CAGAA__GGA___CUG_C__G ...............(((((.......)))))..(((....((....))((..((((((.....))))))..))....)))............................. (-21.34 = -21.28 + -0.05)

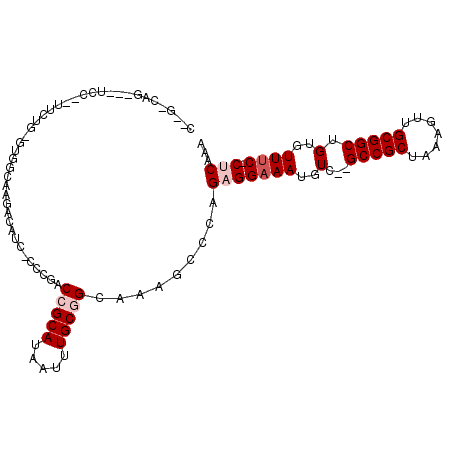
| Location | 6,170,731 – 6,170,835 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6170731 104 - 20766785 CCUGUCAGAGUUCC--UUCUG-GUGGCAAGACGUC-CCCGACCGCAUAAUUUGCGGCAAAGCCCUGAGGAAAUGUC--GCCGCUAAAGUUGCGGCUGUGUUUCCCACCAA ....((((.(((..--....(-(.(((.....)))-.))(.(((((.....))))))..))).))))(((((..(.--(((((.......))))).)..)))))...... ( -36.00) >DroPse_CAF1 7722 96 - 1 -----------UCC--UUCCGUGCCCCAAGACAUGGCCCCACCGCAUAAUUUGCAGCAAAGCCCAGAGGAAAUGUCCCGCCGCUAAAGUUGCGGCUGUGUUUC-CUCAAA -----------...--..(((((........))))).......(((.....)))...........(((((((..(...(((((.......))))).)..))))-)))... ( -28.40) >DroEre_CAF1 12003 105 - 1 CAAGGCGGUGUUCCCAGUCUG-GAGGCAAGACGCC-CCCGACCGCAUAAUUUGCGGCAAAGCCCCGAGGAAAUGUC--GCCGCUAAAGUUGCGGCUGUGUUUC-CACCAA ...(((..(((.....(((.(-(.(((.....)))-)).)))((((.....)))))))..)))....(((((..(.--(((((.......))))).)..))))-)..... ( -42.50) >DroYak_CAF1 8983 101 - 1 CCAGGCAGU--UCC--UUCUG-GUGGCAAGACGUC-CCCGACCGCAUAAUUUGCGGCAAAGCCCCGAGGAAAUGUC--GCCGCUAAAGUUGCGGCUGUGUUUC-CUCGAA ...(((...--...--....(-(.(((.....)))-.))(.(((((.....))))))...))).((((((((..(.--(((((.......))))).)..))))-)))).. ( -43.10) >DroAna_CAF1 12180 103 - 1 CUGGAGACAGCUCC--UUUUG-GUGGCAAGACAUC-CCCGACCGCAUAAUUUGCGGCAAUGCCCGGAGGAAAUGUC--GCCGCUAAAGUUGCGGCUGUGUUGC-CUCAAA ..((((....))))--.((((-(.(((((.(((((-((((...(((.....)))(((...)))))).)))......--(((((.......)))))))).))))-)))))) ( -41.60) >DroPer_CAF1 7744 96 - 1 -----------UCC--UUCCGUACCCCAAGACAUGGCCCCACCGCAUAAUUUGCAGCAAAGCCCAGAGGAAAUGUCCCGCCGCUAAAGUUGCGGCUGUGUUUC-CUCAAA -----------...--..................(((....(.(((.....))).)....)))..(((((((..(...(((((.......))))).)..))))-)))... ( -27.00) >consensus C__G_CAG___UCC__UUCUG_GUGGCAAGACAUC_CCCGACCGCAUAAUUUGCGGCAAAGCCCAGAGGAAAUGUC__GCCGCUAAAGUUGCGGCUGUGUUUC_CUCAAA .........................................(((((.....))))).........(((((((..(...(((((.......))))).)..)))).)))... (-19.88 = -20.72 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:26 2006