
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,163,361 – 6,163,454 |
| Length | 93 |
| Max. P | 0.956851 |

| Location | 6,163,361 – 6,163,454 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6163361 93 + 20766785 ---------UACCUUUUAACUU-------AAAUUUAUUUAGCUCUUCCCAAAGGAGCUUCCGUGCCUUCUUUGGGAUUAUCCAACAAGGCUGUCUACAAGGUAGAUUCU- ---------((((((.((((..-------..........(((((((.....))))))).....(((((..((((......)))).))))).)).)).))))))......- ( -22.00) >DroSec_CAF1 1607 93 + 1 ---------CACCUUUUAACUU-------AAAUUUAUUUAGCUCUUCCCAAAGGAGCUCUCGUGCCUUCUUUGGGAUUAUCCAACAAGGCUGUCUACAAGGUAGAAUCU- ---------.............-------.........((((.((((((((((((((......).)))))))))))..........))))))(((((...)))))....- ( -21.40) >DroSim_CAF1 1606 93 + 1 ---------UACCCUUUAACUU-------AAAUUUAUUUAGCUCUUCCAAAAGGAGCUCUCGUGCCUUCUUUGGGAUUAUCCAACAAGGCUGUCUACAAGGUAGAAUCU- ---------((((.........-------..........(((((((.....))))))).....(((((..((((......)))).))))).........))))......- ( -19.90) >DroEre_CAF1 1608 97 + 1 ---------UAGCUUUUAACUU---AUUAAACUUUAUUUAGCUCUUCCCAAAGGAGCUUCCGUGCCUUCUUUGGGAUUAUCCAACAAGGCUAUCUACAAGGUGGAAUCU- ---------(((((((......---.(((((.....)))))....((((((((((((......).))))))))))).........)))))))(((((...)))))....- ( -23.30) >DroYak_CAF1 1610 106 + 1 UAGUUUACUUACCUUUUAAUUU---AUUUAACUUUAUAUAGCUCUUCCCAAAGGAGCUUCCGUGCCUUCUUUGGGAUUAUCCAACAAGGCUAUCUACAAAGUCGAAUCA- ......................---(((..(((((..(((((.((((((((((((((......).)))))))))))..........)))))))....)))))..)))..- ( -23.70) >DroAna_CAF1 1598 101 + 1 ---------UUCCUUUUUAUAUAAAAUUUGUCUUGUUUUAGCUCUUCCGAAAGGCGCUUCUGUGCCUUCCCUCGGACUGUCGAACAAAGCCGUUUACAAAGAGGAAUCAC ---------(((((((((...((((....(..((((((..((...((((((((((((....)))))))...)))))..)).))))))..)..)))).))))))))).... ( -30.00) >consensus _________UACCUUUUAACUU_______AAAUUUAUUUAGCUCUUCCCAAAGGAGCUUCCGUGCCUUCUUUGGGAUUAUCCAACAAGGCUGUCUACAAGGUAGAAUCU_ ......................................(((((..((((((((((((......).)))))))))))...........)))))(((((...)))))..... (-16.66 = -16.94 + 0.28)

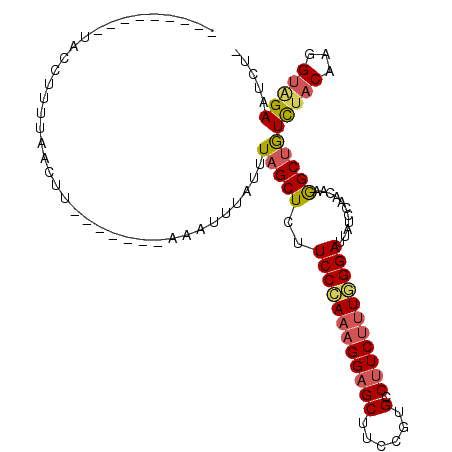
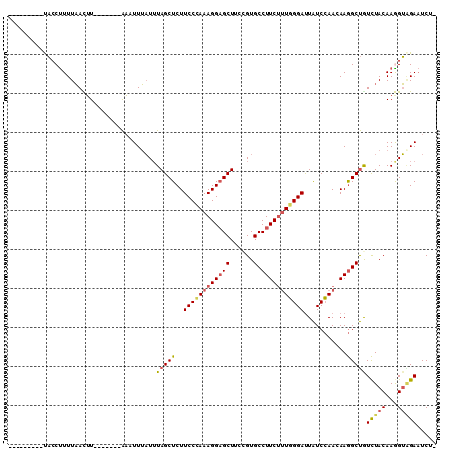
| Location | 6,163,361 – 6,163,454 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -19.53 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6163361 93 - 20766785 -AGAAUCUACCUUGUAGACAGCCUUGUUGGAUAAUCCCAAAGAAGGCACGGAAGCUCCUUUGGGAAGAGCUAAAUAAAUUU-------AAGUUAAAAGGUA--------- -......((((((...(((.(((((.((((......))))..))))).....(((((.........)))))..........-------..)))..))))))--------- ( -24.40) >DroSec_CAF1 1607 93 - 1 -AGAUUCUACCUUGUAGACAGCCUUGUUGGAUAAUCCCAAAGAAGGCACGAGAGCUCCUUUGGGAAGAGCUAAAUAAAUUU-------AAGUUAAAAGGUG--------- -......((((((...(((.(((((.((((......))))..))))).....(((((.........)))))..........-------..)))..))))))--------- ( -24.20) >DroSim_CAF1 1606 93 - 1 -AGAUUCUACCUUGUAGACAGCCUUGUUGGAUAAUCCCAAAGAAGGCACGAGAGCUCCUUUUGGAAGAGCUAAAUAAAUUU-------AAGUUAAAGGGUA--------- -......((((((...(((.(((((.((((......))))..))))).....(((((.........)))))..........-------..)))..))))))--------- ( -24.20) >DroEre_CAF1 1608 97 - 1 -AGAUUCCACCUUGUAGAUAGCCUUGUUGGAUAAUCCCAAAGAAGGCACGGAAGCUCCUUUGGGAAGAGCUAAAUAAAGUUUAAU---AAGUUAAAAGCUA--------- -.................((((((((((......((((((((..(((......))).)))))))).(((((......))))))))---)))......))))--------- ( -20.50) >DroYak_CAF1 1610 106 - 1 -UGAUUCGACUUUGUAGAUAGCCUUGUUGGAUAAUCCCAAAGAAGGCACGGAAGCUCCUUUGGGAAGAGCUAUAUAAAGUUAAAU---AAAUUAAAAGGUAAGUAAACUA -..(((..((((((((....(((((.((((......))))..))))).....(((((.........))))).))))))))..)))---...................... ( -25.70) >DroAna_CAF1 1598 101 - 1 GUGAUUCCUCUUUGUAAACGGCUUUGUUCGACAGUCCGAGGGAAGGCACAGAAGCGCCUUUCGGAAGAGCUAAAACAAGACAAAUUUUAUAUAAAAAGGAA--------- ....(((((.((((((...((((((.((((......))))(((((((.(....).)))))))...)))))).....((((....)))).)))))).)))))--------- ( -24.50) >consensus _AGAUUCUACCUUGUAGACAGCCUUGUUGGAUAAUCCCAAAGAAGGCACGGAAGCUCCUUUGGGAAGAGCUAAAUAAAGUU_______AAGUUAAAAGGUA_________ ........(((((...(((.(((((.((((......))))..))))).....(((((.........)))))...................)))..))))).......... (-19.53 = -18.73 + -0.80)

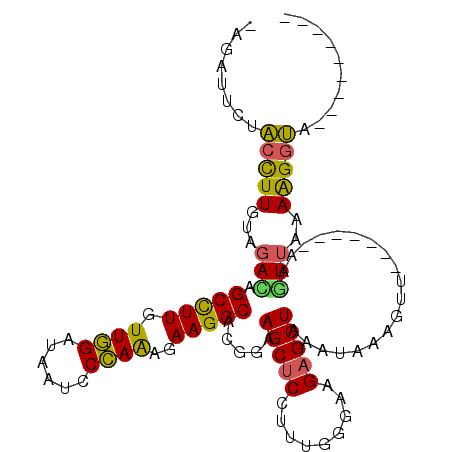
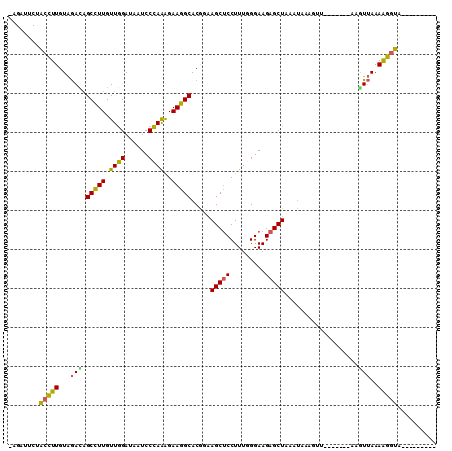
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:23 2006