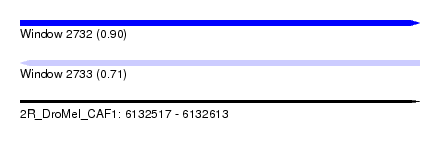
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,132,517 – 6,132,613 |
| Length | 96 |
| Max. P | 0.901599 |

| Location | 6,132,517 – 6,132,613 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -13.53 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
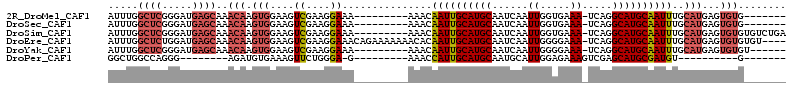
>2R_DroMel_CAF1 6132517 96 + 20766785 -------CACACUCAUGCAAAUUGCAUGCCUGA-UUUCACCAAUUGAUUGCAUGCAAUUGUUU---------UUUCCUUCGACUUCCACUUGUUUGCUCAUCCCGAGCCAAAU -------.........((((.((((((((..((-((.........))))))))))))))))..---------...................((((((((.....))).))))) ( -17.50) >DroSec_CAF1 141595 96 + 1 -------CACACUCAUGCAAAUUGCAUGCCUGA-UUUCACCAAUUGAUUGCAUGCAAUUGUUU---------UUUCCUUCGACUUCCACUUGUUUGCUCAUCCCGAGCCAAAU -------.........((((.((((((((..((-((.........))))))))))))))))..---------...................((((((((.....))).))))) ( -17.50) >DroSim_CAF1 149568 103 + 1 UCAGACACACACUCAUGCAAAUUGCAUGCCUGA-UUUCACCAAUUGAUUGCAUGCAAUUGUUU---------UUUCCUUCGACUUCCACUUGUUUGCUCAUCCCGAGCCAAAU ..(((((............((((((((((..((-((.........))))))))))))))(((.---------........))).......)))))((((.....))))..... ( -17.70) >DroEre_CAF1 145808 108 + 1 ----ACACACACUCAUGCAAAUUGCAUGCCUGA-UUUCCCCAAUUGAUUGCAUGCAAUUGUGUUUUUUUCUGUUUCCUUCGACUUCCACUUGUUUGCUCAUCCAGAGCCAAAU ----..(((.......(((((((((((((..((-((.........)))))))))))))).))).......)))..................((((((((.....))).))))) ( -19.24) >DroYak_CAF1 144934 97 + 1 ------ACACACUCAUGCAAAUUGCAUGCCUGA-UUUCCCCAAUUGAUUGCAUGCAAUUGUUU---------UUUCCUUCGACUUCCACUUGUUUGCUCAUCCCGAGCCAAAU ------..........((((.((((((((..((-((.........))))))))))))))))..---------...................((((((((.....))).))))) ( -17.50) >DroPer_CAF1 134884 78 + 1 -------C----------ACAUCGCAUGCUCGACUUUCUCCAAUGCAUUGCAUGCAAUGGUUU---------C-UCCCAGAACUUUCACAUCU--------CCCUGGCCAGCC -------.----------.....((((((....................))))))...((((.---------.-..((((.............--------..))))..)))) ( -13.61) >consensus _______CACACUCAUGCAAAUUGCAUGCCUGA_UUUCACCAAUUGAUUGCAUGCAAUUGUUU_________UUUCCUUCGACUUCCACUUGUUUGCUCAUCCCGAGCCAAAU ...................((((((((((....................))))))))))....................................((((.....))))..... (-13.53 = -14.08 + 0.56)


| Location | 6,132,517 – 6,132,613 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -15.92 |
| Energy contribution | -17.03 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
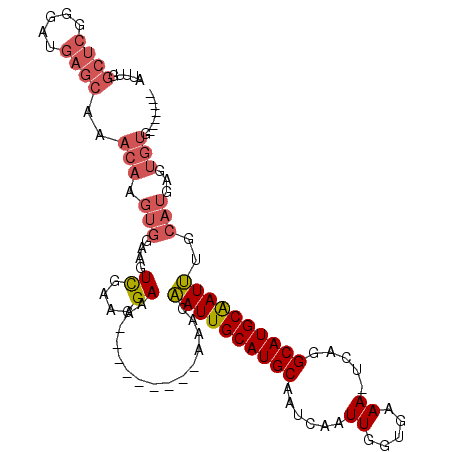
>2R_DroMel_CAF1 6132517 96 - 20766785 AUUUGGCUCGGGAUGAGCAAACAAGUGGAAGUCGAAGGAAA---------AAACAAUUGCAUGCAAUCAAUUGGUGAAA-UCAGGCAUGCAAUUUGCAUGAGUGUG------- .....((((.....))))..(((.(((.((.((....))..---------.....(((((((((..(((.....)))..-....))))))))))).)))...))).------- ( -22.10) >DroSec_CAF1 141595 96 - 1 AUUUGGCUCGGGAUGAGCAAACAAGUGGAAGUCGAAGGAAA---------AAACAAUUGCAUGCAAUCAAUUGGUGAAA-UCAGGCAUGCAAUUUGCAUGAGUGUG------- .....((((.....))))..(((.(((.((.((....))..---------.....(((((((((..(((.....)))..-....))))))))))).)))...))).------- ( -22.10) >DroSim_CAF1 149568 103 - 1 AUUUGGCUCGGGAUGAGCAAACAAGUGGAAGUCGAAGGAAA---------AAACAAUUGCAUGCAAUCAAUUGGUGAAA-UCAGGCAUGCAAUUUGCAUGAGUGUGUGUCUGA .(((.(((...(((..(((..(((.((....((....))..---------...)).)))..))).)))....))).)))-(((((((((((.((.....)).))))))))))) ( -26.10) >DroEre_CAF1 145808 108 - 1 AUUUGGCUCUGGAUGAGCAAACAAGUGGAAGUCGAAGGAAACAGAAAAAAACACAAUUGCAUGCAAUCAAUUGGGGAAA-UCAGGCAUGCAAUUUGCAUGAGUGUGUGU---- .((((((((.....)))).............((....))..)))).....((((((((((((((.....(((.....))-)...)))))))))).(((....)))))))---- ( -24.50) >DroYak_CAF1 144934 97 - 1 AUUUGGCUCGGGAUGAGCAAACAAGUGGAAGUCGAAGGAAA---------AAACAAUUGCAUGCAAUCAAUUGGGGAAA-UCAGGCAUGCAAUUUGCAUGAGUGUGU------ .....((((.....))))..(((.(((.((.((....))..---------.....(((((((((.....(((.....))-)...))))))))))).)))...)))..------ ( -21.70) >DroPer_CAF1 134884 78 - 1 GGCUGGCCAGGG--------AGAUGUGAAAGUUCUGGGA-G---------AAACCAUUGCAUGCAAUGCAUUGGAGAAAGUCGAGCAUGCGAUGU----------G------- ......((((((--------...(.....).))))))..-.---------....((((((((((..((.(((......))))).)))))))))).----------.------- ( -18.60) >consensus AUUUGGCUCGGGAUGAGCAAACAAGUGGAAGUCGAAGGAAA_________AAACAAUUGCAUGCAAUCAAUUGGUGAAA_UCAGGCAUGCAAUUUGCAUGAGUGUG_______ .....((((.....))))..(((.(((....((....))...............((((((((((......((.....)).....))))))))))..)))...)))........ (-15.92 = -17.03 + 1.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:21 2006