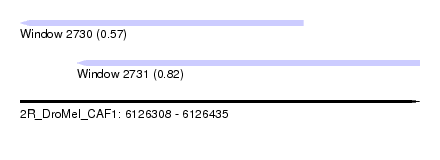
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,126,308 – 6,126,435 |
| Length | 127 |
| Max. P | 0.819966 |

| Location | 6,126,308 – 6,126,398 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.63 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -23.16 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6126308 90 - 20766785 GUUGGCGGAUGCGGCGACGGACUAUACGGGAACUGUUGCUGUAACAUAUGCAACU-UUGCAUAAAUAAAUU-GA-------------UUAUCUCGGUUAGUCAAA- .(((((...(((((((((((.((....))...)))))))))))...((((((...-.))))))....((((-((-------------.....)))))).))))).- ( -26.10) >DroVir_CAF1 161721 88 - 1 GUCGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACU-UUGCAUAAAUAAAUU-UG-------------UUAU--CGGUUAGUCAAA- ((((.((....)).)))).(((((..((..((((((((...)))))((((((...-.))))))........-.)-------------))..--))..)))))...- ( -26.70) >DroPse_CAF1 129030 105 - 1 GUUGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACUGUUGCAUAAAUAAAUCAGAUUGUUUUUAAUUGUUCUAUCGGUUAGUCAAA- .(((((.(((((((((((((.((....))...))))))))))....((((((.....)))))).....)))..........((((((......))))))))))).- ( -29.80) >DroGri_CAF1 153111 89 - 1 GUCGGCGGAUGUGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACU-UUGCAUAAAUAAAUU-UG-------------UUAU--UGGUUAGUCAAGC ...(((...((..(((((((.((....))...)))))))..))...((((((...-.))))))........-..-------------....--......))).... ( -23.60) >DroMoj_CAF1 161469 88 - 1 GUCUGCGCAUGCGGCGACGGACUGUACGGGAACUGAUGCUGCAACAUAUGCAACU-ACGCAUAAAUAAAUU-UG-------------UUAA--CGGUUAGUCAAA- ((((((((.....))).)))))((.((...(((((((((((((.....))))...-..)))).((((....-))-------------))..--))))).))))..- ( -19.70) >DroPer_CAF1 128872 105 - 1 GUUGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACUGUUGCAUAAAUAAAUCAGAUUGUUUUUAAUUGUUCUAUCGGUUAGUCAAA- .(((((.(((((((((((((.((....))...))))))))))....((((((.....)))))).....)))..........((((((......))))))))))).- ( -29.80) >consensus GUCGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACU_UUGCAUAAAUAAAUU_GA_____________UUAU__CGGUUAGUCAAA_ ...(((...(((((((((((.((....))...)))))))))))...((((((.....))))))....................................))).... (-23.16 = -23.38 + 0.22)

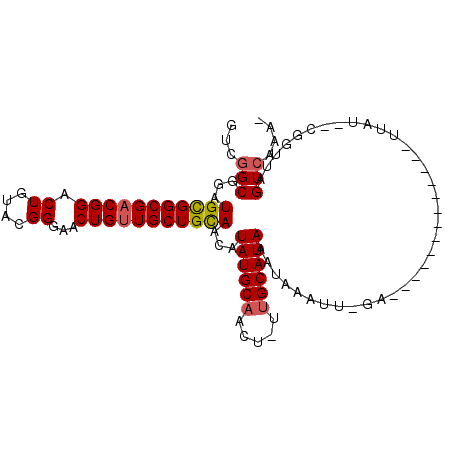
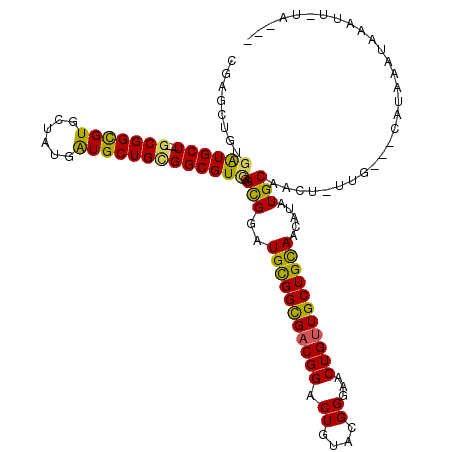
| Location | 6,126,326 – 6,126,435 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -29.20 |
| Energy contribution | -28.32 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6126326 109 - 20766785 CGAGCUGUGGUGCU---GCGGCGUGCUGUGAUGCUGCGGCGUUGGCGGAUGCGGCGACGGACUAUACGGGAACUGUUGCUGUAACAUAUGCAACU-UUG---CAUAAAUAAAUU-GA--- ....((((..((((---(((((((......)))))))))))..).))).(((((((((((.((....))...)))))))))))...((((((...-.))---))))........-..--- ( -41.30) >DroVir_CAF1 161737 109 - 1 CGCGCCAUGGUGCU---GCGGCGUGCCGUGGUGCUGCGGCGUCGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACU-UUG---CAUAAAUAAAUU-UG--- ((((((..((..(.---.(((....)))..)..))..)))).)).....(((((((((((.((....))...)))))))))))...((((((...-.))---))))........-..--- ( -48.30) >DroGri_CAF1 153128 100 - 1 ---------AUGCU---GCGGCGUACUAUGAUGCUGUGGCGUCGGCGGAUGUGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACU-UUG---CAUAAAUAAAUU-UG--- ---------((((.---.((((((......))))))..)))).......((..(((((((.((....))...)))))))..))...((((((...-.))---))))........-..--- ( -35.40) >DroWil_CAF1 149385 119 - 1 GCUGCUGUGAUGCUGUUGCGGUGUACUAUGAUGCUGUGGCGUUGGUGGAUGCGGUGACGGACUGUAUGGAAACUGUUGCUGCAACAUAUGCAACU-UUGGGGUGUAAAUAAAGAAAAAAA (((.(...(.(((.((..((((((......))))))..))....(((..(((((..((((.((....))...))))..))))).)))..))).).-..).)))................. ( -32.00) >DroMoj_CAF1 161485 109 - 1 CGUGCCAUGAUGCU---GCGGCGUUCCGUGAUGCUGCGGCGUCUGCGCAUGCGGCGACGGACUGUACGGGAACUGAUGCUGCAACAUAUGCAACU-ACG---CAUAAAUAAAUU-UG--- .((((...((((((---((((((((....)))))))))))))).)))).(((((((.(((.((....))...))).)))))))...(((((....-..)---))))........-..--- ( -45.70) >DroPer_CAF1 128902 112 - 1 CGAGCUGUGAUGCU---GCGGCGUGCUAUGAUGCUGGGGCGUUGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACUGUUG---CAUAAAUAAAUCAGAU-- ...(((..((((((---.((((((......)))))).))))))))).(((((((((((((.((....))...))))))))))....((((((.....))---)))).....)))....-- ( -42.10) >consensus CGAGCUGUGAUGCU___GCGGCGUGCUAUGAUGCUGCGGCGUCGGCGGAUGCGGCGACGGACUGUACGGGAACUGUUGCUGCAACAUAUGCAACU_UUG___CAUAAAUAAAUU_UA___ ........((((((...(((((((......))))))))))))).(((..(((((((((((.((....))...))))))))))).....)))............................. (-29.20 = -28.32 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:19 2006