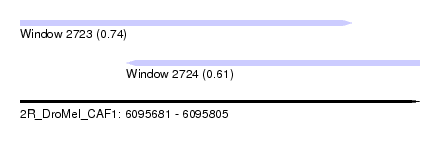
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,095,681 – 6,095,805 |
| Length | 124 |
| Max. P | 0.743042 |

| Location | 6,095,681 – 6,095,784 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6095681 103 + 20766785 AAGAAGUGCAGCAAAAGCAAAUGUACUGAAAGC------GAGGAAUACAUUCUUCUUCUUCAAAAAAAAAUACCAUCAAAAAGAAAGUAAGAAA-----AAGUU-U-UUUCUGCAA ......(((((.((((((........((((...------((((((....))))))...))))........(((..((.....))..))).....-----..)))-)-)).))))). ( -18.90) >DroSec_CAF1 113455 101 + 1 AAGAAGUGCAGCCAAAGCAAAUGUACUGAAAGCAAGAAGGAGGAAUACGUUCUUCU------GAAA-AAAUACCAUCAA-AAGAAAGUAAGAAA-----AAGUU-U-UUACUGCCA ....((((((((....))...))))))....((.(((((((.(....).)))))))------....-............-.....((((((((.-----...))-)-))))))).. ( -18.80) >DroSim_CAF1 118091 103 + 1 AAGAAGUGCAGCCAAAGCAAAUGUACUGAAAGGAAGACGAAGGAAUACGUUCUUCU------GAAAAAAAUACCAUCAA-AAGAAAGUAAGAAA-----AAGUU-UUUUGCUGCAA ......((((((.(((((.............((((((((........)).))))))------........(((..((..-..))..))).....-----..)))-))..)))))). ( -20.00) >DroEre_CAF1 117306 109 + 1 AAGAAGUGCAGCAAAAGCAAAUGUACUGAAAGCAACACGGAGGAAUACAUUCUUCU------GAAAGAAAUACCAGCAU-AAGAAAUCAAGAAAGAGAAAAGUUAUUUUCGUACAA ......(((.......)))..(((((.(((((.(((.((((((((....)))))))------)................-......((......)).....))).)))))))))). ( -19.40) >DroYak_CAF1 115326 101 + 1 AAGAAGUGCAGCAAAAGCAAAUGUACUGAAAGCAAGAAUGAUGAAUGCGUUCUUCU------GAAA-AAAUACCAUCAC-AAGAAA---AGAAAGA---AAGUU-UUUUCGCAUAA ....((((((((....))...))))))....((.....(((((......(((....------))).-......))))).-..((((---(((....---...))-))))))).... ( -16.82) >consensus AAGAAGUGCAGCAAAAGCAAAUGUACUGAAAGCAAGAAGGAGGAAUACGUUCUUCU______GAAA_AAAUACCAUCAA_AAGAAAGUAAGAAA_____AAGUU_UUUUCCUGCAA ....((((((((....))...))))))............((((((....))))))............................................................. ( -8.52 = -8.92 + 0.40)


| Location | 6,095,714 – 6,095,805 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -11.96 |
| Consensus MFE | -0.08 |
| Energy contribution | -0.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.01 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6095714 91 - 20766785 UCUUUUUUUACUUAAGCUCUCUUGCAGAAA-A-AACUU-----UUUCUUACUUUCUUUUUGAUGGUAUUUUUUUUUGAAGAAGAAGAAUGUAUUCCUC--- (((((((((......((......))(((((-(-....)-----)))))((((.((.....)).))))...........)))))))))...........--- ( -12.60) >DroSec_CAF1 113491 84 - 1 UU--UUUUUCCUUAAGCUUUCUGGCAGUAA-A-AACUU-----UUUCUUACUUUCUU-UUGAUGGUAUUU-UUUC------AGAAGAACGUAUUCCUCCUU ..--...........(((....))).....-.-.....-----.....(((.(((((-((((..(....)-..))------))))))).)))......... ( -12.80) >DroSim_CAF1 118127 87 - 1 UCU-UUUUUACUUAAGCUUGCUUGCAGCAAAA-AACUU-----UUUCUUACUUUCUU-UUGAUGGUAUUUUUUUC------AGAAGAACGUAUUCCUUCGU ..(-(((((.((((((....)))).)).))))-))...-----.....(((.(((((-((((..(.....)..))------))))))).)))......... ( -14.10) >DroEre_CAF1 117342 93 - 1 UCU-UUUUUACUUAAGCUUUCUUGUACGAAAAUAACUUUUCUCUUUCUUGAUUUCUU-AUGCUGGUAUUUCUUUC------AGAAGAAUGUAUUCCUCCGU ...-...........((.(((((....(((((....)))))................-...((((........))------))))))).)).......... ( -8.00) >DroYak_CAF1 115362 85 - 1 UCU-UUCUUACUUAAGCUUUCUUAUGCGAAAA-AACUU---UCUUUCU---UUUCUU-GUGAUGGUAUUU-UUUC------AGAAGAACGCAUUCAUCAUU ...-...........((.(((((.((.(((((-.(((.---((.....---......-..)).))).)))-)).)------).))))).)).......... ( -12.32) >consensus UCU_UUUUUACUUAAGCUUUCUUGCAGGAAAA_AACUU_____UUUCUUACUUUCUU_UUGAUGGUAUUU_UUUC______AGAAGAACGUAUUCCUCCGU ...............((......))............................................................................ ( -0.08 = -0.04 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:12 2006