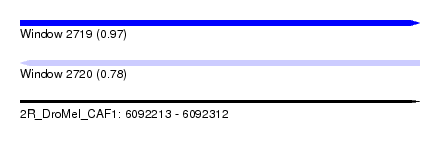
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,092,213 – 6,092,312 |
| Length | 99 |
| Max. P | 0.971255 |

| Location | 6,092,213 – 6,092,312 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6092213 99 + 20766785 AAAGCCAACAAAUGAAAUGUAAGUUUCACUCAUCGAUGGCCAGCAGCAG---UGGAUACGACUAAAAUUGAAUCCAAUAUGAGCCGCCAAGCUGCUGAGCCA ............((((((....))))))........(((((((((((..---(((((.(((......))).)))))...((......)).))))))).)))) ( -28.30) >DroSec_CAF1 109950 93 + 1 AAAGCCAACAAAUGAACUGUAAGUUUCACUCAUCGAUGGCCAGCAGC------GGAUACGACUAAAAUUGAAUCCAAUAUGAGC---CAAGCUGCUGAGCCA .((((..(((.......)))..))))..........(((((((((((------((((.(((......))).))))....((...---)).))))))).)))) ( -23.60) >DroEre_CAF1 113446 99 + 1 AAAGCCAACAAAUGAACUGUAAGUUUCACUCAGCGAUGGCCAGCAGCAGCGGCGGAUACGACUAAAAUUGAAUCCAAUAUGAGC---CAAGCUGCUGAGCCA ...((........((((.....))))......))..(((((((((((...(((((((.(((......))).)))).......))---)..))))))).)))) ( -27.45) >DroYak_CAF1 111710 93 + 1 AAAGCCAACAAAUGAACUGUAAGUUUCACUCAUCGAUGGCCAGCAG------CGGAUACGACUAAAAUUGAAUCCAAUAUGAGC---CAAGCUGCUGAGCCA .((((..(((.......)))..))))..........((((((((((------(((((.(((......))).))))....((...---)).))))))).)))) ( -23.60) >consensus AAAGCCAACAAAUGAACUGUAAGUUUCACUCAUCGAUGGCCAGCAGC_____CGGAUACGACUAAAAUUGAAUCCAAUAUGAGC___CAAGCUGCUGAGCCA .((((..(((.......)))..))))..........(((((((((((......((((.(((......))).))))....((......)).))))))).)))) (-22.48 = -22.72 + 0.25)

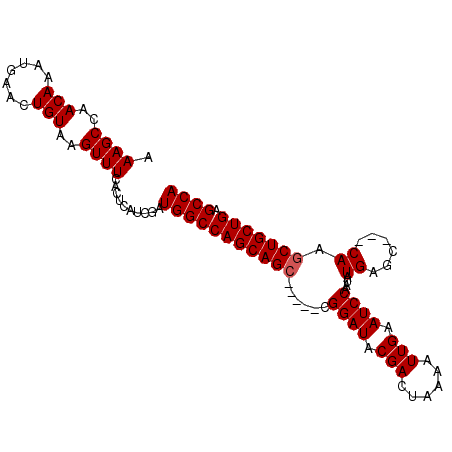
| Location | 6,092,213 – 6,092,312 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -26.51 |
| Energy contribution | -27.01 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.93 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6092213 99 - 20766785 UGGCUCAGCAGCUUGGCGGCUCAUAUUGGAUUCAAUUUUAGUCGUAUCCA---CUGCUGCUGGCCAUCGAUGAGUGAAACUUACAUUUCAUUUGUUGGCUUU ((((.(((((((.((((((((...((((....))))...)))))...)))---..)))))))))))((((..(((((((......)))))))..)))).... ( -39.20) >DroSec_CAF1 109950 93 - 1 UGGCUCAGCAGCUUG---GCUCAUAUUGGAUUCAAUUUUAGUCGUAUCC------GCUGCUGGCCAUCGAUGAGUGAAACUUACAGUUCAUUUGUUGGCUUU ((((.(((((((.((---(((...((((....))))...))))).....------)))))))))))((((..((((((........))))))..)))).... ( -33.20) >DroEre_CAF1 113446 99 - 1 UGGCUCAGCAGCUUG---GCUCAUAUUGGAUUCAAUUUUAGUCGUAUCCGCCGCUGCUGCUGGCCAUCGCUGAGUGAAACUUACAGUUCAUUUGUUGGCUUU ((((.(((((((.((---((..((((..((((.......))))))))..))))..)))))))))))..((((((((((........)))))))...)))... ( -32.70) >DroYak_CAF1 111710 93 - 1 UGGCUCAGCAGCUUG---GCUCAUAUUGGAUUCAAUUUUAGUCGUAUCCG------CUGCUGGCCAUCGAUGAGUGAAACUUACAGUUCAUUUGUUGGCUUU ((((.(((((((.((---(((...((((....))))...))))).....)------))))))))))((((..((((((........))))))..)))).... ( -33.20) >consensus UGGCUCAGCAGCUUG___GCUCAUAUUGGAUUCAAUUUUAGUCGUAUCCG_____GCUGCUGGCCAUCGAUGAGUGAAACUUACAGUUCAUUUGUUGGCUUU ((((.(((((((............((((((......)))))).............)))))))))))((((((((((((........)))))))))))).... (-26.51 = -27.01 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:09 2006