
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,072,377 – 6,072,519 |
| Length | 142 |
| Max. P | 0.818034 |

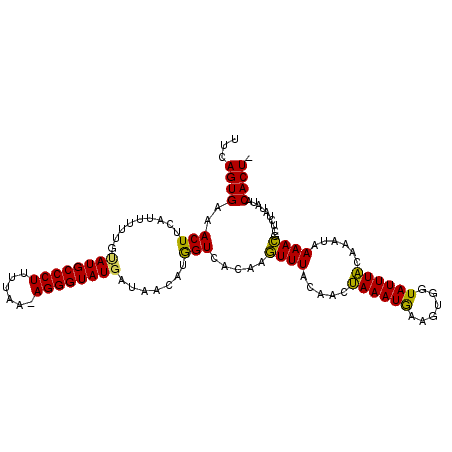
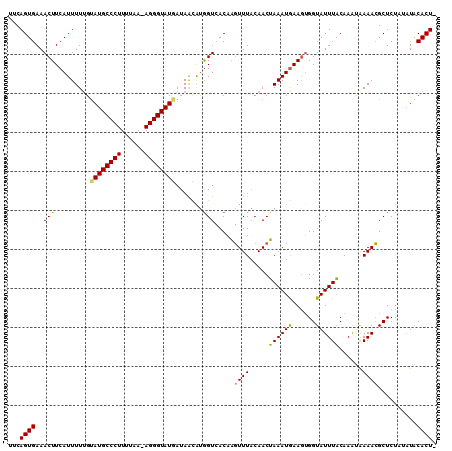
| Location | 6,072,377 – 6,072,486 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -14.76 |
| Energy contribution | -13.66 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6072377 109 - 20766785 UUCAGUGAAACUUCAUUUUUGUAUGCCCUUUUAAAAGGGUAUCAUAGUAUGGUCACAUGUUUACAACAAAAUAAA-UGGUAUUUUCAAAUAAAACGCUCUUUUUACACUA ...((((..(((....((((((((((((((....)))))))).....((((....))))......))))))....-.))).........(((((......))))))))). ( -17.80) >DroEre_CAF1 92734 108 - 1 UUCAGUGAAACUUCAUUUUUGUAUGCCCUUUCAA-AGGGUAUGAUAACGCGGUCACAAGUUUUCAACUAAAUGAAGUGGUAUUUACAAGUAAAACGCUCUAAAUACACU- .........(((((((((..(((((((((.....-)))))))((.(((..(....)..))).)).)).))))))))).(((((((..(((.....))).)))))))...- ( -24.50) >DroYak_CAF1 91573 109 - 1 UUCAGUGAAACUUCAUUUUCGCAUGCCCUUUUAA-AGGGUAUGAUAACAUAGUAACUAUUUUACAACUAAAUGAAGAGGUAUUUACAAACGAAAAGCUCUAUAUGCACUC ....((((((((((.......((((((((.....-))))))))........((((.....))))...........))))).))))).........((.......)).... ( -22.70) >consensus UUCAGUGAAACUUCAUUUUUGUAUGCCCUUUUAA_AGGGUAUGAUAACAUGGUCACAAGUUUACAACUAAAUGAAGUGGUAUUUACAAAUAAAACGCUCUAUAUACACU_ ...((((..(((.........((((((((......)))))))).......))).....((((.....((((((......))))))......))))..........)))). (-14.76 = -13.66 + -1.10)

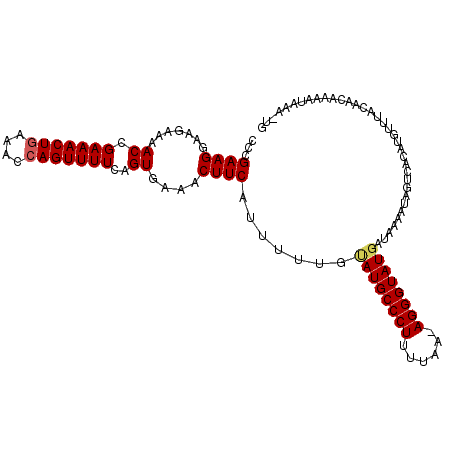
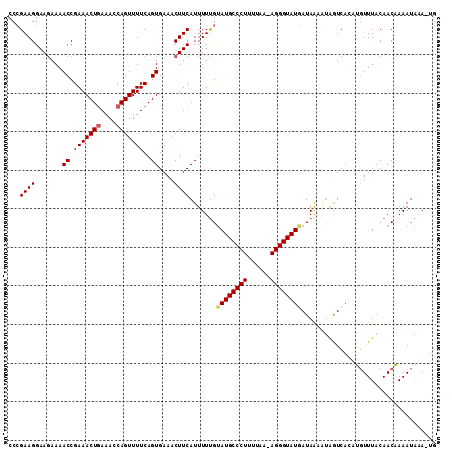
| Location | 6,072,409 – 6,072,519 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.10 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -17.82 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6072409 110 - 20766785 CCAGAAGGAAGAAAACCGAAACUGAAACCAGUUUUCAGUGAAACUUCAUUUUUGUAUGCCCUUUUAAAAGGGUAUCAUAGUAUGGUCACAUGUUUACAACAAAAUAAA-UG .......((((...((.(((((((....)))))))..))....))))..((((((((((((((....)))))))).....((((....))))......))))))....-.. ( -22.60) >DroSec_CAF1 90607 109 - 1 CCCGAAGGAAGAAAACCGAAACUAAAACCAGUUUUCAGUGAAACUUCAUUUUUGUAUGCCCUUUUAA-AGGGUAUGAUAAAAUAAUCAGAUGUUUACAACAAAAUAUA-UG ...((..((((...((.((((((......))))))..))....))))..(((((((((((((.....-))))))).))))))...))..((((((......)))))).-.. ( -18.60) >DroSim_CAF1 95521 109 - 1 CCCGAAGGAAGAAAACCGAAACUGAAACCAGUUUUCAGUGAAACUUCAUUUUUGUAUGCCCUUUUAA-AGGGUAUGAUAAAAUAAUCAGAUGUUUACAACAAAAUAUA-UG ...((..((((...((.(((((((....)))))))..))....))))..(((((((((((((.....-))))))).))))))...))..((((((......)))))).-.. ( -21.70) >DroEre_CAF1 92765 110 - 1 CCCGAAGGAAGAAAACCGAAACUGAAACCAGUUUUCAGUGAAACUUCAUUUUUGUAUGCCCUUUCAA-AGGGUAUGAUAACGCGGUCACAAGUUUUCAACUAAAUGAAGUG ......((.......))...(((((((.....)))))))...(((((((((..(((((((((.....-)))))))((.(((..(....)..))).)).)).))))))))). ( -25.60) >DroYak_CAF1 91605 109 - 1 CCCGAAGGAA-AAAACCGAAACUGAAACCAGUUUUCAGUGAAACUUCAUUUUCGCAUGCCCUUUUAA-AGGGUAUGAUAACAUAGUAACUAUUUUACAACUAAAUGAAGAG ......((..-....))...(((((((.....)))))))....((((((((...((((((((.....-))))))))........((((.....))))....)))))))).. ( -26.10) >consensus CCCGAAGGAAGAAAACCGAAACUGAAACCAGUUUUCAGUGAAACUUCAUUUUUGUAUGCCCUUUUAA_AGGGUAUGAUAAAAUAGUCACAUGUUUACAACAAAAUAAA_UG ...((((.......((.(((((((....)))))))..))....)))).......((((((((......))))))))................................... (-17.82 = -18.10 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:02 2006