
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,063,169 – 6,063,266 |
| Length | 97 |
| Max. P | 0.642548 |

| Location | 6,063,169 – 6,063,266 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6063169 97 - 20766785 --------AAGCAUUACUUUUCGCUGAUGCGCCCUCGGGGCAAAUCG--------GUGAAGCGCCCACACCCAUCUCCAAAAAUGUACAAAAACAGCCAACACACACUCACAC --------..((......((((((((((..((((...))))..))))--------))))))......................(((......)))))................ ( -19.70) >DroSec_CAF1 81534 85 - 1 --------AAGCAUUACUUUUCGCUGAUGCGCCCUCGGGGCAAAUCG--------UUAAAGCGCCCACACCCAUCUCCAAAAAUGUACAAAAACACCCAAC------------ --------..(((((......(((....))).....(((......((--------(....)))......))).........)))))...............------------ ( -12.60) >DroSim_CAF1 85848 97 - 1 --------AAGCAUUACUUUUCGCUGAUGCGCCCUCGGGGCAAAUCG--------GUGAAGCGCCCACACCCAUCUCCAAAAAUGUACAAAAACACCCAACACACAUUCACAC --------..(((((...((((((((((..((((...))))..))))--------)))))).(....).............)))))........................... ( -18.50) >DroEre_CAF1 83231 97 - 1 --------AAGCAUUACUUUUCGCUGAAGCGCCCUCGGGGCGAAUCG--------GUGAAGCGCCCACACCCAUCUCCGAAAAUGUACAAAAACACCCAGCACACACUCACAC --------..((......(((((((((..(((((...)))))..)))--------))))))......................(((......)))....))............ ( -22.20) >DroYak_CAF1 82209 95 - 1 AAGCUUCAAAGCAUUACUUUUCGCUGAUGCGCCCUCGGGGCGAAUCG--------GUGAAGCGCCCACACCCAUCUCCAAAAAUGUACAA----------CACACACACGCAC ..(((....))).......(((((((((.(((((...))))).))))--------)))))(((...........................----------........))).. ( -23.35) >DroPer_CAF1 79832 89 - 1 --------AAGCAUUACUUUUCGCUGAUGCGCCCUCGGGGCGAGUCAGUCAGUCAGUCAGGCGCCCACACCCAUCUCCAUACAUGUAGGUA--CACCUC-------------- --------..((...(((.((.((((((.(((((...))))).)))))).))..)))...))(((.(((..............))).))).--......-------------- ( -22.54) >consensus ________AAGCAUUACUUUUCGCUGAUGCGCCCUCGGGGCAAAUCG________GUGAAGCGCCCACACCCAUCUCCAAAAAUGUACAAAAACACCCAACACACACUCACAC ..........((.((((.....((....))((((...))))..............)))).))................................................... (-12.11 = -12.33 + 0.22)

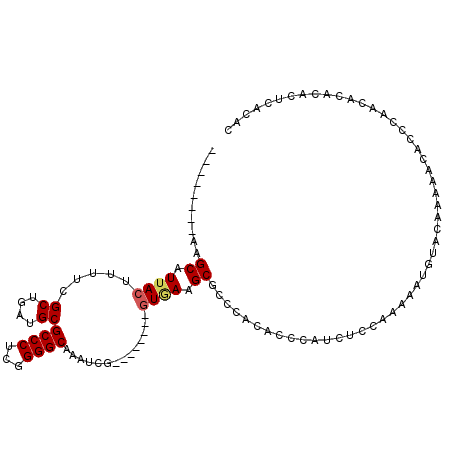
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:00 2006