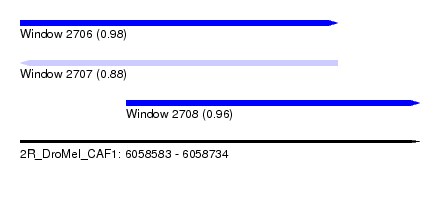
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,058,583 – 6,058,734 |
| Length | 151 |
| Max. P | 0.975595 |

| Location | 6,058,583 – 6,058,703 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -16.54 |
| Energy contribution | -18.26 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6058583 120 + 20766785 CACACUAGUAUGUACAUAUGUACUAUGUAUGUGAAGCCAUUGCCUUGGCAAAACAAAGCAACAAAAUUCGCCAUACUUUAGCUAGGCCCAAAAAUAGCCAAAUCCUGGGAAAACUCCCAC ....(((((..((((....))))...(((((((((....((((.(((......))).)))).....)))).)))))....)))))....................(((((....))))). ( -28.30) >DroSec_CAF1 77051 112 + 1 CUCA--------UACACAUGUGCUAUGUAUGUGGACCAACUGCUUGGGCAAAACAAAGCAACAAAAUUCGCCAUACUUUAGCUAGGCCCGAAAAUAGCCAAAUCCUGGGAAAACUCCCAC ....--------.........((((.(((((((((.....(((((.(......).)))))......)))).)))))..))))..(((.........)))......(((((....))))). ( -24.80) >DroSim_CAF1 80807 112 + 1 CUCA--------UACACAUGUGCUAUGUAUGUGGACCAACUGCUUGGGCAAAACAAAGCAACACAAUUCGCCAUACUUUAGCUAGGCCCGAAAAUAGCCAAAUCCUGGGAAAACUCCCAC ....--------.........((((.(((((((((.....(((((.(......).)))))......)))).)))))..))))..(((.........)))......(((((....))))). ( -24.80) >DroEre_CAF1 78651 98 + 1 ------------UAUGUAUGG---AUGCU--UUGUACAUAUGCUUUGG-----CAAAGCAACAAAAUUCGCUAUACUUAAGCUAGGCCCGAAAGUGGCCAAGUCCUGGGAAAACUACCAA ------------...((((((---.((((--((((.((.......)))-----)))))))..........))))))....(((.(((((....).)))).)))..(((........))). ( -27.80) >DroYak_CAF1 77581 105 + 1 ------------UACAUAUGU---AUGUAUGUGGACCAACUGCUUUGGUAAAACAAAGCAACAAAAUUUGCUAUACCUAAGCUAGGCCCGAAAAUAGCCAAGUCCUGGGAAAACUCCCAU ------------.((((((..---..))))))((((.....((((.((((......(((((......))))).)))).))))..(((.........)))..))))(((((....))))). ( -29.00) >consensus C_CA________UACAUAUGU_CUAUGUAUGUGGACCAACUGCUUUGGCAAAACAAAGCAACAAAAUUCGCCAUACUUUAGCUAGGCCCGAAAAUAGCCAAAUCCUGGGAAAACUCCCAC ..........................(((((((((.....(((((((......)))))))......))))).))))........(((.........)))......(((((....))))). (-16.54 = -18.26 + 1.72)



| Location | 6,058,583 – 6,058,703 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.94 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6058583 120 - 20766785 GUGGGAGUUUUCCCAGGAUUUGGCUAUUUUUGGGCCUAGCUAAAGUAUGGCGAAUUUUGUUGCUUUGUUUUGCCAAGGCAAUGGCUUCACAUACAUAGUACAUAUGUACAUACUAGUGUG .((((......))))......((((.......))))..((((..(((((..(((..(..((((((((......))))))))..).))).)))))...((((....))))....))))... ( -34.40) >DroSec_CAF1 77051 112 - 1 GUGGGAGUUUUCCCAGGAUUUGGCUAUUUUCGGGCCUAGCUAAAGUAUGGCGAAUUUUGUUGCUUUGUUUUGCCCAAGCAGUUGGUCCACAUACAUAGCACAUGUGUA--------UGAG .((((......))))(((.(..(((.(((..((((..(((.((((((..((((...)))))))))))))..))))))).)))..)))).((((((((.....))))))--------)).. ( -36.20) >DroSim_CAF1 80807 112 - 1 GUGGGAGUUUUCCCAGGAUUUGGCUAUUUUCGGGCCUAGCUAAAGUAUGGCGAAUUGUGUUGCUUUGUUUUGCCCAAGCAGUUGGUCCACAUACAUAGCACAUGUGUA--------UGAG .((((......))))(((.(..(((.(((..((((..(((.((((((..((.....))..)))))))))..))))))).)))..)))).((((((((.....))))))--------)).. ( -37.00) >DroEre_CAF1 78651 98 - 1 UUGGUAGUUUUCCCAGGACUUGGCCACUUUCGGGCCUAGCUUAAGUAUAGCGAAUUUUGUUGCUUUG-----CCAAAGCAUAUGUACAA--AGCAU---CCAUACAUA------------ .(((........)))(((...((((.......))))..((((..(((.(((((......))))).))-----)..))))..........--....)---)).......------------ ( -22.80) >DroYak_CAF1 77581 105 - 1 AUGGGAGUUUUCCCAGGACUUGGCUAUUUUCGGGCCUAGCUUAGGUAUAGCAAAUUUUGUUGCUUUGUUUUACCAAAGCAGUUGGUCCACAUACAU---ACAUAUGUA------------ .((((......))))(((((.((((.......))))..((((.((((.((((((.........)))))).)))).))))....)))))(((((...---...))))).------------ ( -33.10) >consensus GUGGGAGUUUUCCCAGGAUUUGGCUAUUUUCGGGCCUAGCUAAAGUAUGGCGAAUUUUGUUGCUUUGUUUUGCCAAAGCAGUUGGUCCACAUACAUAG_ACAUAUGUA________UG_G (((((........((((((((((((.......))))..((((.....))))))))))))((((((((......))))))))....))))).((((((.....))))))............ (-20.46 = -21.94 + 1.48)


| Location | 6,058,623 – 6,058,734 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.86 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6058623 111 + 20766785 UGCCUUGGCAAAACAAAGCAACAAAAUUCGCCAUACUUUAGCUAGGCCCAAAAAUAGCCAAAUCCUGGGAAAACUCCCACACUCCC----CC----UUUUUUCUGGCAAGA-ACAAGGAC ..(((((((........))..........((((...........(((.........)))......(((((....))))).......----..----.......))))....-.))))).. ( -23.60) >DroSec_CAF1 77083 110 + 1 UGCUUGGGCAAAACAAAGCAACAAAAUUCGCCAUACUUUAGCUAGGCCCGAAAAUAGCCAAAUCCUGGGAAAACUCCCACACCUCC----CC----UU-UUUCUGGCCAGA-ACAAGGAC (((((.(......).)))))...............((((..((.((((.(((((...........(((((....))))).......----..----.)-)))).)))))).-..)))).. ( -24.60) >DroSim_CAF1 80839 110 + 1 UGCUUGGGCAAAACAAAGCAACACAAUUCGCCAUACUUUAGCUAGGCCCGAAAAUAGCCAAAUCCUGGGAAAACUCCCACACCGCC----CC----UU-UUUCUGGCCAGC-ACAAGGAC ..(((((((....................)))........(((.((((.(((((..((.......(((((....)))))....)).----..----.)-)))).)))))))-.))))... ( -28.45) >DroEre_CAF1 78674 111 + 1 UGCUUUGG-----CAAAGCAACAAAAUUCGCUAUACUUAAGCUAGGCCCGAAAGUGGCCAAGUCCUGGGAAAACUACCAACCCCCCCGCACCCCCAUU-UCACUCCCCUUUUCCAAG--- ....((((-----.((((......................(((.(((((....).)))).)))..((((.......................))))..-........)))).)))).--- ( -21.20) >DroYak_CAF1 77606 111 + 1 UGCUUUGGUAAAACAAAGCAACAAAAUUUGCUAUACCUAAGCUAGGCCCGAAAAUAGCCAAGUCCUGGGAAAACUCCCAUCCCCCCUGCUUC----UU-UUACUGGCA----GCAAGGAC .((((.((((......(((((......))))).)))).))))..(((.........)))..(((((((((....)))).......(((((..----..-.....))))----)..))))) ( -30.20) >consensus UGCUUUGGCAAAACAAAGCAACAAAAUUCGCCAUACUUUAGCUAGGCCCGAAAAUAGCCAAAUCCUGGGAAAACUCCCACACCCCC____CC____UU_UUUCUGGCCAGA_ACAAGGAC (((((((......))))))).........((((...........(((.........)))......(((((....)))))........................))))............. (-17.78 = -18.86 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:58 2006