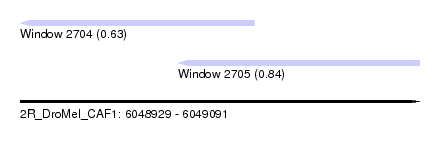
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,048,929 – 6,049,091 |
| Length | 162 |
| Max. P | 0.842309 |

| Location | 6,048,929 – 6,049,024 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -17.07 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
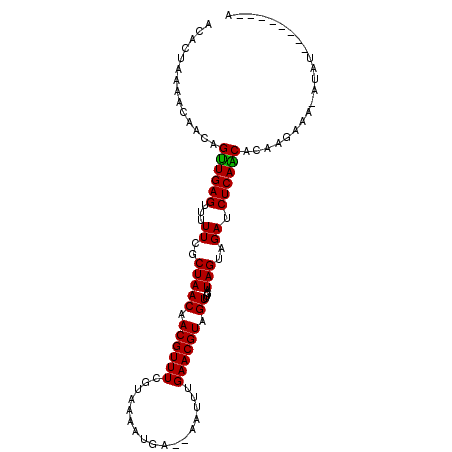
>2R_DroMel_CAF1 6048929 95 - 20766785 AUAUUAAAACAA--GUUGAGUUUUUCGCUAACAACGUUUCGUAAAAUGAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAAGGAA-AUAUG-------A ............--((((((...((..(((((.((((((...(((((....))))))))))).))....)))..)).))))))........-.....-------. ( -18.20) >DroPse_CAF1 66906 92 - 1 ACACCAAAAAUGCAGUUGAGUUUUUCGCUAACAACGUUUUGUAUAAUGA--AAUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAGUAU-----------UCUC ........(((((.((((((...((..(((((.((((((...((.....--.))..)))))).))....)))..)).))))))...))))-----------)... ( -16.00) >DroEre_CAF1 68076 97 - 1 AUAUUAAAACAAAUGUUGAGUUUUUCGCUAACAACGUUUCGUAAAAUGAUCAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAAGAAA-AUAUU-------A .............(((((((...((..(((((.((((((...(((((....))))))))))).))....)))..)).))))))).......-.....-------. ( -17.00) >DroYak_CAF1 66881 96 - 1 AGAUUAAAACAAAAGUUGAGUUUUUCGCUAACAACGUUUCGUAA-AUGAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAAGGAA-AUAUU-------A ..............((((((...((..(((((.((((((...((-((....)))).)))))).))....)))..)).))))))........-.....-------. ( -18.10) >DroAna_CAF1 70307 104 - 1 AAACUAAUCCA-CAGCUGAGUUUUUCGCUAACAACGUUUCUUAAAAUGAUUAAUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAGUCACUAUAUCAUCUUUUA ...........-..((((((...((..(((((.((((((....((((.....)))))))))).))....)))..)).))))))...................... ( -17.80) >DroPer_CAF1 66603 92 - 1 ACACCAAAAAUGCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAAUGA--AAUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAGUAU-----------UCUC ........(((((.((((((...((..(((.....(((((((...))))--)))(..(((...)))..))))..)).))))))...))))-----------)... ( -15.30) >consensus ACACUAAAACAACAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAAUGA__AAUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAAGAAA_AUAU________A ..............((((((...((..(((((.((((((.................)))))).))....)))..)).))))))...................... (-15.06 = -14.70 + -0.36)

| Location | 6,048,993 – 6,049,091 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
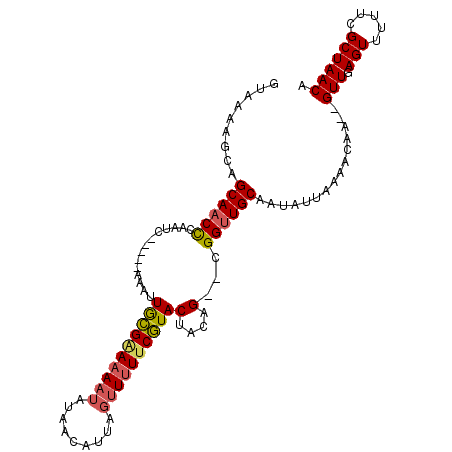
>2R_DroMel_CAF1 6048993 98 - 20766785 AUUAAAGCAGCAACCCAAUC-----AAAUUGCGAAAAAUAUAACAUUAGUUUUUCGUACUACAG--CGGUUGCAAUAUUAAAACAA--GUUGAGUUUUUCGCUAACA .....(((.((((((.....-----....((((((((((.........))))))))))......--.))))))......(((((..--.....)))))..))).... ( -20.19) >DroPse_CAF1 66965 101 - 1 --CCAUUCAGCAUCUCGUUCAUCGUAAAGUAUGAAAAUUAGUACA----CUUUCCAUACGACAGAACGGUUGCAACACCAAAAAUGCAGUUGAGUUUUUCGCUAACA --..(((((((...((((((.(((((((((.((........)).)----)))....)))))..)))))).((((..........)))))))))))............ ( -18.30) >DroSec_CAF1 67128 98 - 1 GUAAAAGCAGCAACCCAAUC-----AAAUUGCGAAAAAUAUAACAUUAGUUUUUCGUACUACAG--CGGUUGCAAUAUUAACCCAA--GUUGAGUUUUUCGCUAACA ((((((((.((((((.....-----....((((((((((.........))))))))))......--.))))))....(((((....--))))))))))).))..... ( -21.89) >DroSim_CAF1 70382 97 - 1 GUAAAAGCAGCAACCUAAUC-----AAAUUGCGAAAAAUAUAACAUUAGUU-UUCGUACUACAG--CGGUUGCAAUAUUAACCCAA--GUUGAGUUUUUCGCUAACA ((((((((.((((((.....-----....((((((((.((......)).))-))))))......--.))))))....(((((....--))))))))))).))..... ( -20.79) >DroEre_CAF1 68140 97 - 1 GUAAAA---GCAACCCAAUC-----AAAUUGCGGAAAAUAUAACAUCAGUUUUUCGUACUACAG--UGGUUGCAAUAUUAAAACAAAUGUUGAGUUUUUCGCUAACA ((((((---((....(((((-----(...((((((((((.........))))))))))......--))))))(((((((......))))))).)))))).))..... ( -19.20) >DroYak_CAF1 66944 97 - 1 GUAAGA---GCAACCCAAUC-----AAAUUGCGGAAAAUAUACCAUCAGUUUUUCGUACUACAG--CGGUUGCGAGAUUAAAACAAAAGUUGAGUUUUUCGCUAACA ((.((.---((((((.....-----....((((((((((.........))))))))))......--.))))))((((....(((....))).....)))).)).)). ( -19.79) >consensus GUAAAAGCAGCAACCCAAUC_____AAAUUGCGAAAAAUAUAACAUUAGUUUUUCGUACUACAG__CGGUUGCAAUAUUAAAACAA__GUUGAGUUUUUCGCUAACA .........((((((..............((((((((((.........))))))))))(....)...))))))...............(((.(((.....)))))). (-13.44 = -13.55 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:55 2006