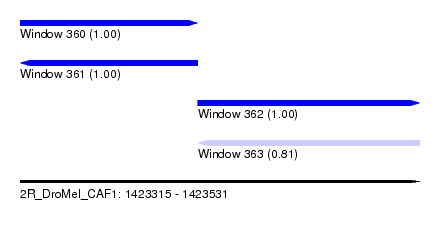
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,423,315 – 1,423,531 |
| Length | 216 |
| Max. P | 0.999909 |

| Location | 1,423,315 – 1,423,411 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -38.76 |
| Consensus MFE | -29.39 |
| Energy contribution | -31.70 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.76 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1423315 96 + 20766785 AAGAGGGGCGGCUGUGAACCCCUCUUGGGGUUCAUGUUCAUACUAAUCAGCCACAGCCAGGCGCAGGCAAACCGAAUGAUCCCUCGGCUCCGCCCC-------------------- ....((((((((((((((((((....)))))))).((....))....)))))..(((((((..((..(.....)..))...))).))))..)))))-------------------- ( -42.40) >DroSec_CAF1 41767 106 + 1 AAGAGGGGCGGCUGUGAACCCCUCUUGGGGUUCAUGUUCAUACUAAUCAGCCACAGCCAGGCGCAGGCAAACCCAAUGAUCCCUGGGCUCCGCCCAAGCUCCC----------AUC ..(.((((((((((((((((((....)))))))..((....)).........)))))).((((.((.....((((........)))))).))))...))))))----------... ( -42.80) >DroSim_CAF1 46817 116 + 1 AAGAGGGGCGGCUGUGAACCCCUCUUGGGGUUCAUGUUCAUACUAAUCAGCCACAGCCAGGCGCAGGCAAACCCAAUGAUCCCUGGGCUCCGCCCCAGCUCCCACCAGCUCCCACC ....((((((((((((((((((....)))))))..((....)).........)))))).(((.((((..............)))).)))..)))))((((......))))...... ( -46.34) >DroYak_CAF1 41130 77 + 1 AA-------------GAACCCCUCUUGGGGUUCAUGUUUAUACUAAUCAGCAACAGCCAGGCAU------ACCCGAUGAUCCCUGGGCUCCGCACC-------------------- ..-------------(((((((....)))))))................((...((((((((((------.....)))...))).))))..))...-------------------- ( -23.50) >consensus AAGAGGGGCGGCUGUGAACCCCUCUUGGGGUUCAUGUUCAUACUAAUCAGCCACAGCCAGGCGCAGGCAAACCCAAUGAUCCCUGGGCUCCGCCCC____________________ ....((((((((((((((((((....)))))))................(((.......))))))).....((((........))))..))))))).................... (-29.39 = -31.70 + 2.31)

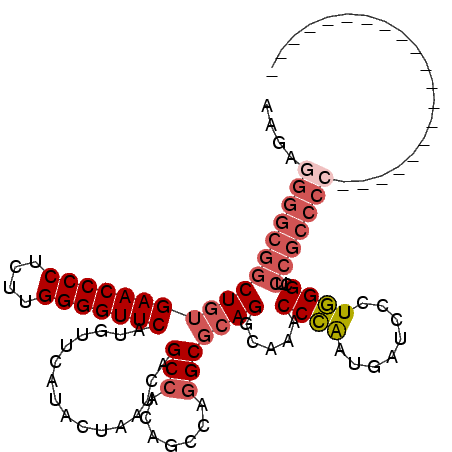
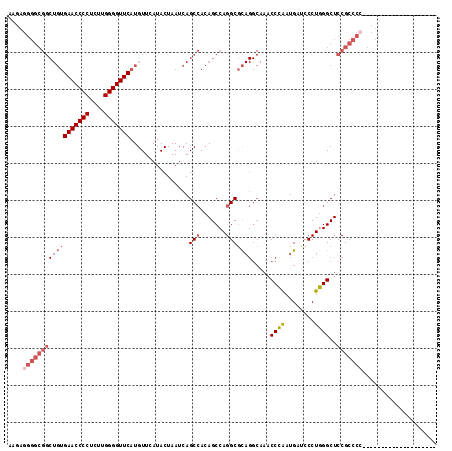
| Location | 1,423,315 – 1,423,411 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -45.25 |
| Consensus MFE | -31.01 |
| Energy contribution | -33.45 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1423315 96 - 20766785 --------------------GGGGCGGAGCCGAGGGAUCAUUCGGUUUGCCUGCGCCUGGCUGUGGCUGAUUAGUAUGAACAUGAACCCCAAGAGGGGUUCACAGCCGCCCCUCUU --------------------(((((.((((((((......))))))))(((.......)))...(((((....((....)).((((((((....)))))))))))))))))).... ( -47.00) >DroSec_CAF1 41767 106 - 1 GAU----------GGGAGCUUGGGCGGAGCCCAGGGAUCAUUGGGUUUGCCUGCGCCUGGCUGUGGCUGAUUAGUAUGAACAUGAACCCCAAGAGGGGUUCACAGCCGCCCCUCUU ((.----------(((((((.(((((((((((((......)))))))...)))).)).))))(((((((....((....)).((((((((....)))))))))))))))))))).. ( -51.60) >DroSim_CAF1 46817 116 - 1 GGUGGGAGCUGGUGGGAGCUGGGGCGGAGCCCAGGGAUCAUUGGGUUUGCCUGCGCCUGGCUGUGGCUGAUUAGUAUGAACAUGAACCCCAAGAGGGGUUCACAGCCGCCCCUCUU ((.(((((((...(((.((..(((((.(((((((......)))))))))))))).)))))))(((((((....((....)).((((((((....)))))))))))))))))).)). ( -55.90) >DroYak_CAF1 41130 77 - 1 --------------------GGUGCGGAGCCCAGGGAUCAUCGGGU------AUGCCUGGCUGUUGCUGAUUAGUAUAAACAUGAACCCCAAGAGGGGUUC-------------UU --------------------.((((.((...(((.(((..(((((.------...)))))..))).))).)).))))......(((((((....)))))))-------------.. ( -26.50) >consensus ____________________GGGGCGGAGCCCAGGGAUCAUUGGGUUUGCCUGCGCCUGGCUGUGGCUGAUUAGUAUGAACAUGAACCCCAAGAGGGGUUCACAGCCGCCCCUCUU .....................(((((((((((((......)))))))).....)))))....(((((((....((....)).((((((((....)))))))))))))))....... (-31.01 = -33.45 + 2.44)

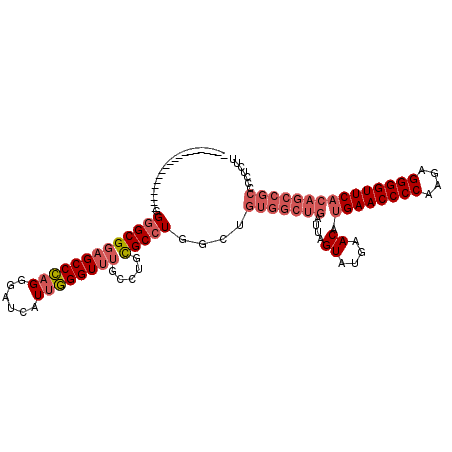
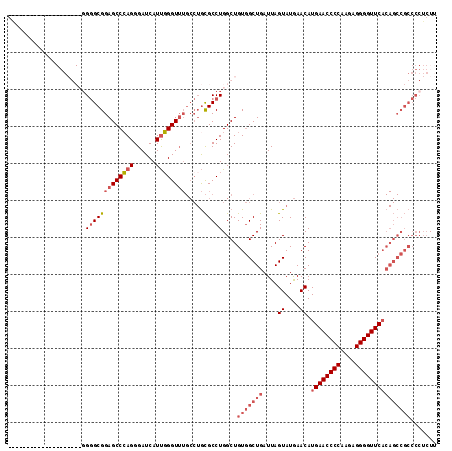
| Location | 1,423,411 – 1,423,531 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -32.06 |
| Energy contribution | -32.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1423411 120 + 20766785 AGCUCCUACCAGCUUGUUUUUGAUUAUAAAUUGAAAAUUUGCUGUUGUGCAAAAUUUGUGCGAAAUCAAAGUUUUUGCGGGGAGAUUCGUUUUUCUUUCGGACAACGGGUUUCUCGGACC ...(((...((((..(((((..((.....))..)))))..))))...(((((((((((........))))..)))))))((((((((((((.(((....))).))))))))))))))).. ( -34.80) >DroSec_CAF1 41873 120 + 1 AGCUCCUACCAGCUUGUUUUUGAUUAUAAAUUGAAAAUUUGCUGUUGUGCGAAAUUUGUGCGAAAUCAAAGUUUUUGCGGGGAGAUUCGUUUUCCUUUCGGACAACGGGUUUCUCGGACC ...(((...((((..(((((..((.....))..)))))..))))...(((((((((((........))))..)))))))((((((((((((.(((....))).))))))))))))))).. ( -37.40) >DroSim_CAF1 46933 119 + 1 AGCUCCUACCAGCUUGUUUUUGAUCAUAAAUUGAAAAUUUGCUGUUGUGCAAAAUUUGUGCAAAAUCAAAGUGUUUGCGGGGAGAUUC-UUUUCCUUUCGGACAACGGGUUUCUCGGACC ...(((...((((..(((((..((.....))..)))))..))))(((..((.....))..)))((((....((((((..((((((...-.))))))..))))))...))))....))).. ( -33.50) >DroEre_CAF1 40055 113 + 1 ---UC---CCAGCUUGUUUUUGAUUAUAAAUUGAAAAUUUGCUGUUGUGCAAAAUUUGUGCGCAAUCAAAGUGUGUGCGGGGAGAUUCGUU-UCCUUUCGGACAACGGGUUUCUCGGACC ---((---(((((..(((((..((.....))..)))))..)))).((..((...((((........))))...))..))((((((((((((-(((....))).))))))))))))))).. ( -37.40) >DroYak_CAF1 41207 119 + 1 AGCUCCU-ACAGCUUGUUUUUGAUUACAAAUUGAAAAUUUGCCGUUUUGCAAAAGUUGUACGAAAUCAAAGUGUUUGCGGGGAGAUUCGUUUUUCUUUCGGAUAACGGGUUUCUCGAACC ...((.(-((((((((((((..((.....))..)))))((((......)))))))))))).))................((((((((((((.(((....))).))))))))))))..... ( -29.80) >consensus AGCUCCUACCAGCUUGUUUUUGAUUAUAAAUUGAAAAUUUGCUGUUGUGCAAAAUUUGUGCGAAAUCAAAGUGUUUGCGGGGAGAUUCGUUUUCCUUUCGGACAACGGGUUUCUCGGACC .........((((..(((((..((.....))..)))))..))))...((((((.((((........))))...))))))((((((((((((.(((....))).))))))))))))..... (-32.06 = -32.46 + 0.40)

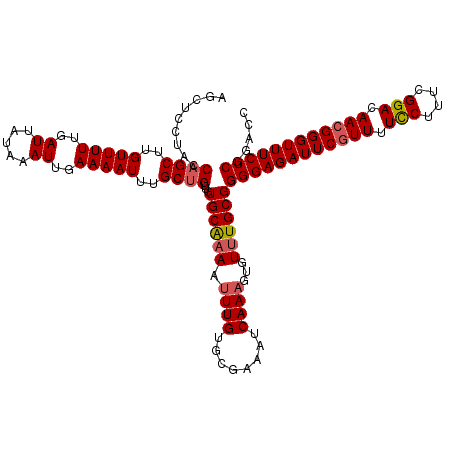
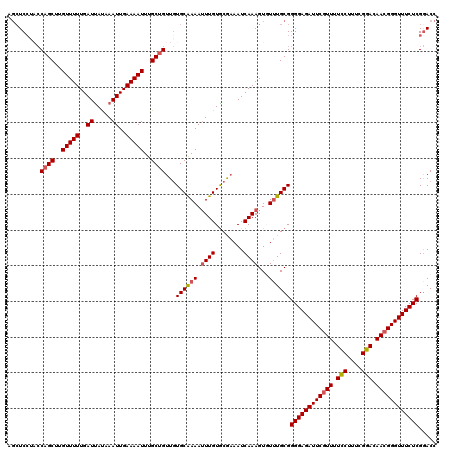
| Location | 1,423,411 – 1,423,531 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.81 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1423411 120 - 20766785 GGUCCGAGAAACCCGUUGUCCGAAAGAAAAACGAAUCUCCCCGCAAAAACUUUGAUUUCGCACAAAUUUUGCACAACAGCAAAUUUUCAAUUUAUAAUCAAAAACAAGCUGGUAGGAGCU (.(((((((....((((...(....)...))))..))))((.((......(((((((......(((.(((((......))))).)))........))))))).....)).))..))).). ( -24.14) >DroSec_CAF1 41873 120 - 1 GGUCCGAGAAACCCGUUGUCCGAAAGGAAAACGAAUCUCCCCGCAAAAACUUUGAUUUCGCACAAAUUUCGCACAACAGCAAAUUUUCAAUUUAUAAUCAAAAACAAGCUGGUAGGAGCU (.(((((((....((((.(((....))).))))..))))((.((......(((((((......((((((.((......)))))))).........))))))).....)).))..))).). ( -27.66) >DroSim_CAF1 46933 119 - 1 GGUCCGAGAAACCCGUUGUCCGAAAGGAAAA-GAAUCUCCCCGCAAACACUUUGAUUUUGCACAAAUUUUGCACAACAGCAAAUUUUCAAUUUAUGAUCAAAAACAAGCUGGUAGGAGCU (.(((((((.........(((....)))...-...))))((.((......(((((((......(((.(((((......))))).)))........))))))).....)).))..))).). ( -25.30) >DroEre_CAF1 40055 113 - 1 GGUCCGAGAAACCCGUUGUCCGAAAGGA-AACGAAUCUCCCCGCACACACUUUGAUUGCGCACAAAUUUUGCACAACAGCAAAUUUUCAAUUUAUAAUCAAAAACAAGCUGG---GA--- .....((((....((((.(((....)))-))))..))))(((((......((((((((.....(((.(((((......))))).))).......)))))))).....)).))---).--- ( -26.60) >DroYak_CAF1 41207 119 - 1 GGUUCGAGAAACCCGUUAUCCGAAAGAAAAACGAAUCUCCCCGCAAACACUUUGAUUUCGUACAACUUUUGCAAAACGGCAAAUUUUCAAUUUGUAAUCAAAAACAAGCUGU-AGGAGCU ((((((((((..(((((...((((((....(((((((................)).)))))....))))))...)))))....)))))..(((((........)))))....-..))))) ( -20.99) >consensus GGUCCGAGAAACCCGUUGUCCGAAAGGAAAACGAAUCUCCCCGCAAACACUUUGAUUUCGCACAAAUUUUGCACAACAGCAAAUUUUCAAUUUAUAAUCAAAAACAAGCUGGUAGGAGCU (.(((((((....((((.(((....))).))))..))))((.((......(((((((..........(((((......)))))............))))))).....)).))..))).). (-20.73 = -21.81 + 1.08)

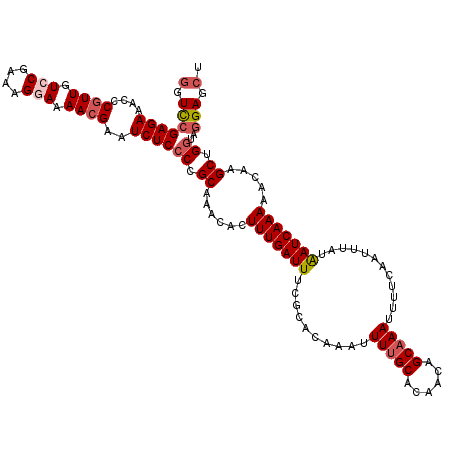
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:38 2006