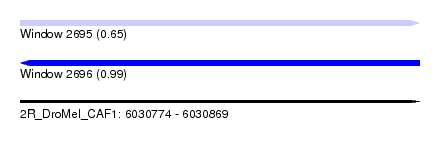
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,030,774 – 6,030,869 |
| Length | 95 |
| Max. P | 0.988650 |

| Location | 6,030,774 – 6,030,869 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 67.10 |
| Mean single sequence MFE | -16.92 |
| Consensus MFE | -5.69 |
| Energy contribution | -5.58 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
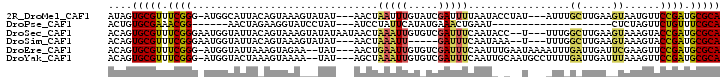
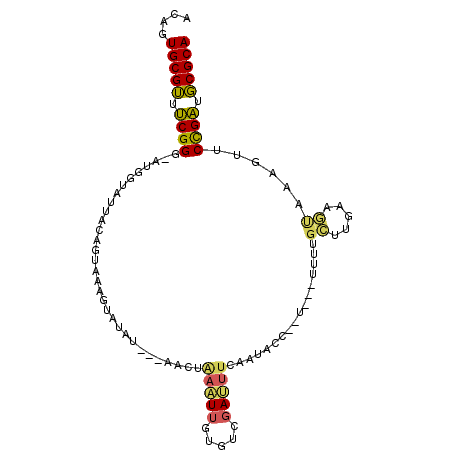
>2R_DroMel_CAF1 6030774 95 + 20766785 UGCGCAUCGGAACAUUACUUCAAGCAAAU---AUAGGUAUUAAAAUCGAUACAAAUUAGUU---AUAUACUUUACUGUAAUGCCAU-CCCGAAACGCACUAU ((((..((((..((((((...........---....(((((......))))).....(((.---....))).....))))))....-.))))..)))).... ( -16.40) >DroPse_CAF1 47692 73 + 1 UGCGAAACAGAACUAGAG--------------------AUUCAGUUUCAUAUGAAUAGGAU---AUAGGAUACCUUCUAGUU------CCGUUUCGCACAGU ((((((((.(((((((((--------------------(((((........))))).((((---.....)).))))))))))------).)))))))).... ( -25.70) >DroSec_CAF1 49022 97 + 1 UGCGCAUCGGUACUUUACUUCAAGCCAAA---A--GGUAUUGAAAUCGACACAAUUUAGUUAUUAUAUACUUUACUGUAAUACCAUUCCCGAAACGCACUGU ((((..((((........((((((((...---.--))).)))))..............(.(((((((........))))))).)....))))..)))).... ( -18.10) >DroSim_CAF1 48749 89 + 1 UGCGCAUCGGUACUUUACUUCAAGCCAAA---A--UUUAUUGAAAUC-----AAUUUAGUU---AUAUACUUUACUGUAAUACCAUUCCCGAAACGCACUGU ((((..((((........(((((......---.--....)))))...-----......(((---(((........)))))).......))))..)))).... ( -12.80) >DroEre_CAF1 49281 96 + 1 UGCGCAUCGGAACUUCGAAUCAAUCAAAUUUUAUUCAAAUUGAAAUCGACACAAUUCAGUU---AUA--UUCUACUUUAAUACCAU-CCCGAAACGCACUGU ((((..((((......((((.((......)).)))).(((((((..........)))))))---...--.................-.))))..)))).... ( -14.80) >DroYak_CAF1 48586 96 + 1 UGCGCAUCGGAACUUUAAAUCAAUCAAAAGGCAUUGCAAUUGAAAUCGACACAAUUUAGCU---AUA--UUUUACUUUAGUACCAU-CCCGAAACGCACUGU ((((..((((.............((((...((...))..))))...............(((---(..--........)))).....-.))))..)))).... ( -13.70) >consensus UGCGCAUCGGAACUUUACUUCAAGCAAAA___A__GGUAUUGAAAUCGACACAAUUUAGUU___AUAUACUUUACUGUAAUACCAU_CCCGAAACGCACUGU ((((..((((..............................................................................))))..)))).... ( -5.69 = -5.58 + -0.11)

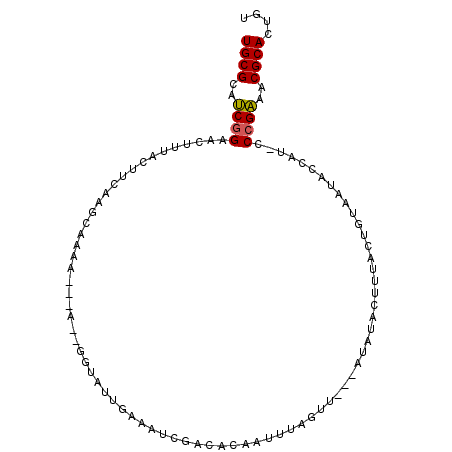
| Location | 6,030,774 – 6,030,869 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 67.10 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.15 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6030774 95 - 20766785 AUAGUGCGUUUCGGG-AUGGCAUUACAGUAAAGUAUAU---AACUAAUUUGUAUCGAUUUUAAUACCUAU---AUUUGCUUGAAGUAAUGUUCCGAUGCGCA ....(((((.(((((-...(((((((....(((((.((---(........((((........))))...)---)).)))))...)))))))))))).))))) ( -22.40) >DroPse_CAF1 47692 73 - 1 ACUGUGCGAAACGG------AACUAGAAGGUAUCCUAU---AUCCUAUUCAUAUGAAACUGAAU--------------------CUCUAGUUCUGUUUCGCA ....((((((((((------((((((((((........---..)))(((((........)))))--------------------.))))))))))))))))) ( -28.60) >DroSec_CAF1 49022 97 - 1 ACAGUGCGUUUCGGGAAUGGUAUUACAGUAAAGUAUAUAAUAACUAAAUUGUGUCGAUUUCAAUACC--U---UUUGGCUUGAAGUAAAGUACCGAUGCGCA ....(((((.(((((((.((((((..(((...(.(((((((......)))))))).)))..))))))--.---))).((((......)))).)))).))))) ( -24.80) >DroSim_CAF1 48749 89 - 1 ACAGUGCGUUUCGGGAAUGGUAUUACAGUAAAGUAUAU---AACUAAAUU-----GAUUUCAAUAAA--U---UUUGGCUUGAAGUAAAGUACCGAUGCGCA ....(((((.((((...........((((..(((....---.)))..)))-----)(((((((....--.---......)))))))......)))).))))) ( -18.40) >DroEre_CAF1 49281 96 - 1 ACAGUGCGUUUCGGG-AUGGUAUUAAAGUAGAA--UAU---AACUGAAUUGUGUCGAUUUCAAUUUGAAUAAAAUUUGAUUGAUUCGAAGUUCCGAUGCGCA ....(((((.(((((-((.(((((.......))--)))---.......(((.(((((((..(((((.....))))).))))))).))).))))))).))))) ( -24.10) >DroYak_CAF1 48586 96 - 1 ACAGUGCGUUUCGGG-AUGGUACUAAAGUAAAA--UAU---AGCUAAAUUGUGUCGAUUUCAAUUGCAAUGCCUUUUGAUUGAUUUAAAGUUCCGAUGCGCA ....(((((.(((((-(((..((...(((....--...---.))).....))..)....(((((..(((......))))))))......))))))).))))) ( -21.10) >consensus ACAGUGCGUUUCGGG_AUGGUAUUACAGUAAAGUAUAU___AACUAAAUUGUGUCGAUUUCAAUACC__U___UUUUGCUUGAAGUAAAGUUCCGAUGCGCA ....(((((.((((...............................(((((.....))))).................((.....))......)))).))))) ( -9.62 = -9.15 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:46 2006