
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,026,148 – 6,026,284 |
| Length | 136 |
| Max. P | 0.999962 |

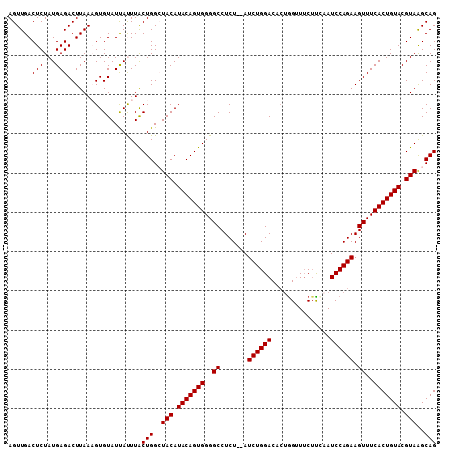
| Location | 6,026,148 – 6,026,257 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6026148 109 + 20766785 AGUUGACUCUAUGAGACUUAAAGUGUAUUAUUUACUGGCUACAUACAGUGGGGCCUCUAUAUCUGGACACUGGUUUCAUUCAUCCAGAAGUUUCACUGUACGUAAGCAG ..((((.((.....)).))))((((.......)))).(((((.(((((((..((.......((((((...(((......))))))))).))..))))))).)).))).. ( -26.70) >DroSec_CAF1 44500 107 + 1 AGUUGACUCUAUGAGACUUAACGUGUAUCAUUUACUGGCUACAUACAGUGGGGCCUCU--AUCUGGACACUGGCUUCUUCAAUCCAGAAGUUUCACUGUACGUAAGCAG .(((((.((.....)).)))))..(((.....)))..(((((.(((((((..((....--.((((((...(((.....))).)))))).))..))))))).)).))).. ( -29.80) >DroSim_CAF1 44235 107 + 1 AGUUGACUCUAUGAGACUUAACGUGUAUCAUUUACUGGCUACAUACAGUGGGGCCUCU--AUCUGGACACGGGUUUCUUCAAUCCAGAAGUUUCACUGUACGUAGGCAG .(((((.((.....)).)))))............(((.((((.(((((((..((....--.((((((....((.....))..)))))).))..))))))).)))).))) ( -35.40) >DroEre_CAF1 44648 106 + 1 AGUUGACUCUAUGAGACUUAAAGUGUAUUACUUGCUGGAUACAUACAGUGGGGCCUCU--AUCUGGACACUGGUUGCUC-AAUCCAGAAGUUUCACUGUACGUAGGCAG ...((.(.(((((.........(((((((........)))))))((((((..((....--.((((((...(((....))-).)))))).))..)))))).)))))))). ( -28.70) >DroYak_CAF1 44133 107 + 1 AGUUGACUCUAUGAGACUUAAAGUGUAUUACUUGCUGGAUACAUACAGUGGGGCCUCU--AUCUGGACAAUAGUUUCUUAAAUCCAGAAGUUUCACUGUACGUAAGCAG (((..((.((.(((...))).)).))...)))((((...(((.(((((((..((....--.((((((....((...))....)))))).))..))))))).))))))). ( -26.30) >consensus AGUUGACUCUAUGAGACUUAAAGUGUAUUAUUUACUGGCUACAUACAGUGGGGCCUCU__AUCUGGACACUGGUUUCUUCAAUCCAGAAGUUUCACUGUACGUAAGCAG ......(((...)))...................(((..(((.(((((((..((.......((((((...............)))))).))..))))))).)))..))) (-23.34 = -23.34 + -0.00)


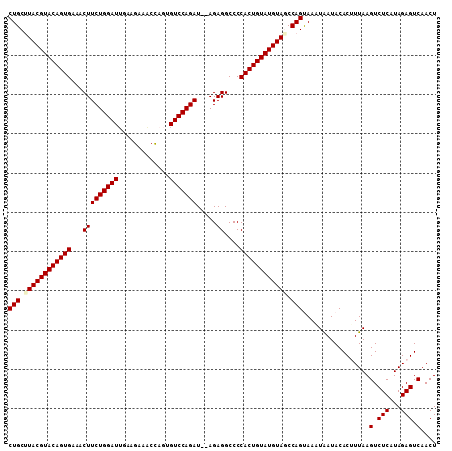
| Location | 6,026,148 – 6,026,257 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6026148 109 - 20766785 CUGCUUACGUACAGUGAAACUUCUGGAUGAAUGAAACCAGUGUCCAGAUAUAGAGGCCCCACUGUAUGUAGCCAGUAAAUAAUACACUUUAAGUCUCAUAGAGUCAACU (((.((((((((((((...(((((((((...((....))..))))))).....))....)))))))))))).)))..........((((((.......))))))..... ( -29.00) >DroSec_CAF1 44500 107 - 1 CUGCUUACGUACAGUGAAACUUCUGGAUUGAAGAAGCCAGUGUCCAGAU--AGAGGCCCCACUGUAUGUAGCCAGUAAAUGAUACACGUUAAGUCUCAUAGAGUCAACU (((.((((((((((((...((((......))))..(((..(((....))--)..)))..)))))))))))).)))...((((.((.......)).)))).......... ( -29.60) >DroSim_CAF1 44235 107 - 1 CUGCCUACGUACAGUGAAACUUCUGGAUUGAAGAAACCCGUGUCCAGAU--AGAGGCCCCACUGUAUGUAGCCAGUAAAUGAUACACGUUAAGUCUCAUAGAGUCAACU (((.((((((((((((...(((((((((.............))))))).--..))....)))))))))))).)))...((((.((.......)).)))).......... ( -31.02) >DroEre_CAF1 44648 106 - 1 CUGCCUACGUACAGUGAAACUUCUGGAUU-GAGCAACCAGUGUCCAGAU--AGAGGCCCCACUGUAUGUAUCCAGCAAGUAAUACACUUUAAGUCUCAUAGAGUCAACU (((..(((((((((((...((((((..((-(..((.....))..))).)--)))))...)))))))))))..)))..........((((((.......))))))..... ( -28.30) >DroYak_CAF1 44133 107 - 1 CUGCUUACGUACAGUGAAACUUCUGGAUUUAAGAAACUAUUGUCCAGAU--AGAGGCCCCACUGUAUGUAUCCAGCAAGUAAUACACUUUAAGUCUCAUAGAGUCAACU (((..(((((((((((...(((((((((.............))))))).--..))....)))))))))))..)))..........((((((.......))))))..... ( -27.92) >consensus CUGCUUACGUACAGUGAAACUUCUGGAUUGAAGAAACCAGUGUCCAGAU__AGAGGCCCCACUGUAUGUAGCCAGUAAAUAAUACACUUUAAGUCUCAUAGAGUCAACU (((.((((((((((((...(((((((((.............))))))).....))....)))))))))))).))).................(.(((...))).).... (-27.00 = -27.32 + 0.32)


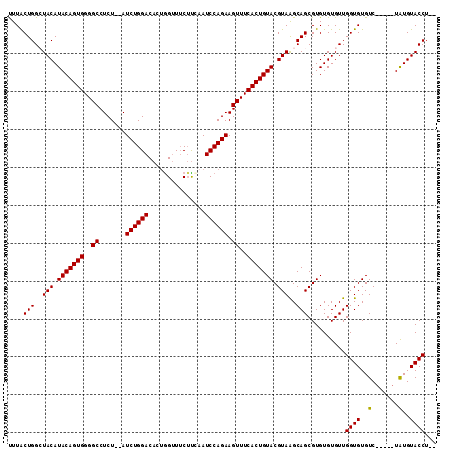
| Location | 6,026,178 – 6,026,284 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6026178 106 + 20766785 UUUACUGGCUACAUACAGUGGGGCCUCUAUAUCUGGACACUGGUUUCAUUCAUCCAGAAGUUUCACUGUACGUAAGCAGCGUGUGUGUUGGUGUGUC-----UAUGUACCU-- ..(((..((.((((((((((..((.......((((((...(((......))))))))).))..))))))((((.....))))))))))..)))....-----.........-- ( -30.20) >DroSec_CAF1 44530 106 + 1 UUUACUGGCUACAUACAGUGGGGCCUCU--AUCUGGACACUGGCUUCUUCAAUCCAGAAGUUUCACUGUACGUAAGCAGCGUGUGUGUUGGUGUGUC-----UAUGUACCUGC ..(((..((.((((((((((..((....--.((((((...(((.....))).)))))).))..))))))((((.....))))))))))..)))....-----........... ( -30.50) >DroSim_CAF1 44265 106 + 1 UUUACUGGCUACAUACAGUGGGGCCUCU--AUCUGGACACGGGUUUCUUCAAUCCAGAAGUUUCACUGUACGUAGGCAGCGUGUGUGUUGGUGUGUC-----UGUGUACCUGC ....(((.((((.(((((((..((....--.((((((....((.....))..)))))).))..))))))).)))).))).......((.((((....-----....)))).)) ( -34.40) >DroEre_CAF1 44678 108 + 1 CUUGCUGGAUACAUACAGUGGGGCCUCU--AUCUGGACACUGGUUGCUC-AAUCCAGAAGUUUCACUGUACGUAGGCAGCGUGUGUGUUGGUGUGUCAUACAUAUGUACCU-- ...((((..(((.(((((((..((....--.((((((...(((....))-).)))))).))..))))))).)))..))))(((((((.........)))))))........-- ( -36.80) >DroYak_CAF1 44163 104 + 1 CUUGCUGGAUACAUACAGUGGGGCCUCU--AUCUGGACAAUAGUUUCUUAAAUCCAGAAGUUUCACUGUACGUAAGCAGCGUGUGUGUUGGUGUGUC-----UGUGUACCU-- ...((((..(((.(((((((..((....--.((((((....((...))....)))))).))..))))))).)))..)))).........((((....-----....)))).-- ( -31.40) >consensus UUUACUGGCUACAUACAGUGGGGCCUCU__AUCUGGACACUGGUUUCUUCAAUCCAGAAGUUUCACUGUACGUAAGCAGCGUGUGUGUUGGUGUGUC_____UAUGUACCU__ ....(((..(((.(((((((..((.......((((((...............)))))).))..))))))).)))..)))..........((((..(.......)..))))... (-27.48 = -27.24 + -0.24)


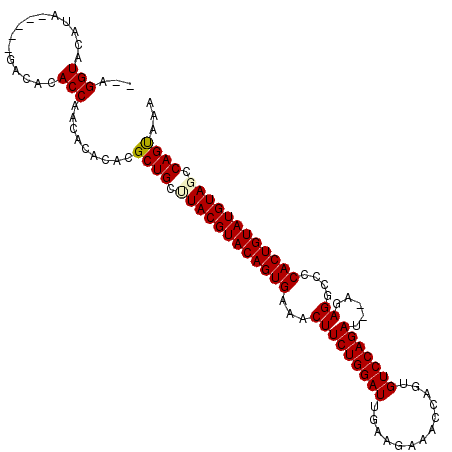
| Location | 6,026,178 – 6,026,284 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.92 |
| SVM RNA-class probability | 0.999962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6026178 106 - 20766785 --AGGUACAUA-----GACACACCAACACACACGCUGCUUACGUACAGUGAAACUUCUGGAUGAAUGAAACCAGUGUCCAGAUAUAGAGGCCCCACUGUAUGUAGCCAGUAAA --.(((.....-----.....))).........((((.((((((((((((...(((((((((...((....))..))))))).....))....)))))))))))).))))... ( -30.80) >DroSec_CAF1 44530 106 - 1 GCAGGUACAUA-----GACACACCAACACACACGCUGCUUACGUACAGUGAAACUUCUGGAUUGAAGAAGCCAGUGUCCAGAU--AGAGGCCCCACUGUAUGUAGCCAGUAAA ...(((.....-----.....))).........((((.((((((((((((...((((......))))..(((..(((....))--)..)))..)))))))))))).))))... ( -31.00) >DroSim_CAF1 44265 106 - 1 GCAGGUACACA-----GACACACCAACACACACGCUGCCUACGUACAGUGAAACUUCUGGAUUGAAGAAACCCGUGUCCAGAU--AGAGGCCCCACUGUAUGUAGCCAGUAAA ...(((.....-----.....))).........((((.((((((((((((...(((((((((.............))))))).--..))....)))))))))))).))))... ( -32.42) >DroEre_CAF1 44678 108 - 1 --AGGUACAUAUGUAUGACACACCAACACACACGCUGCCUACGUACAGUGAAACUUCUGGAUU-GAGCAACCAGUGUCCAGAU--AGAGGCCCCACUGUAUGUAUCCAGCAAG --..((((....)))).................((((..(((((((((((...((((((..((-(..((.....))..))).)--)))))...)))))))))))..))))... ( -33.20) >DroYak_CAF1 44163 104 - 1 --AGGUACACA-----GACACACCAACACACACGCUGCUUACGUACAGUGAAACUUCUGGAUUUAAGAAACUAUUGUCCAGAU--AGAGGCCCCACUGUAUGUAUCCAGCAAG --.(((.....-----.....))).........((((..(((((((((((...(((((((((.............))))))).--..))....)))))))))))..))))... ( -32.32) >consensus __AGGUACAUA_____GACACACCAACACACACGCUGCUUACGUACAGUGAAACUUCUGGAUUGAAGAAACCAGUGUCCAGAU__AGAGGCCCCACUGUAUGUAGCCAGUAAA ...(((...............))).........((((.((((((((((((...(((((((((.............))))))).....))....)))))))))))).))))... (-30.04 = -30.12 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:43 2006