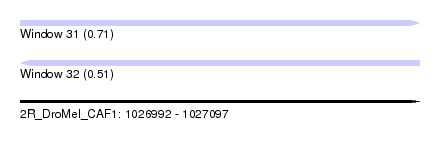
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,026,992 – 1,027,097 |
| Length | 105 |
| Max. P | 0.713295 |

| Location | 1,026,992 – 1,027,097 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -18.17 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
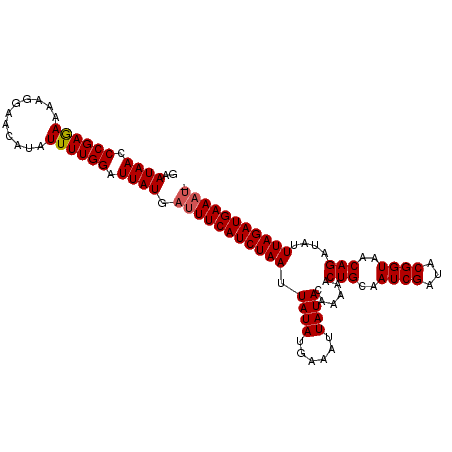
>2R_DroMel_CAF1 1026992 105 + 20766785 GAAUAACCCGAAAAAAGGAACAUAUUUUGGAUUAUGAUUUCAUCUAAUUAUAUGAAAUUAUACAAAAACUGCAAUCGAUACGGUAACAGAUAUUUAGAUGAAAUA ..((((.((((((...........)))))).)))).(((((((((((.(((.((........))....(((..((((...))))..)))))).))))))))))). ( -18.90) >DroSec_CAF1 29442 105 + 1 GAAUAACCCGAGAAAAGGAACAUAUUUUGGAUUAUGAUUUCAUCUAAUUAUAUGAAAUUAUACAAAAACUGCAAUCGAUACGGUAACAGAUAUUUAGAUGAAAUG ..((((.((((((...........)))))).)))).(((((((((((.(((.((........))....(((..((((...))))..)))))).))))))))))). ( -19.00) >DroSim_CAF1 10932 97 + 1 GAAUAACCCGAGAAAAGGAACAUAUUUUGGAUUAUGAUUUCAUCUAAUUAUAUGAAAUUAUA------CUGCAAUCGAUACGGUAACAGAUAUUUAGAUGAAU-- ..((((.((((((...........)))))).))))...(((((((((.((((......))))------(((..((((...))))..)))....))))))))).-- ( -16.60) >consensus GAAUAACCCGAGAAAAGGAACAUAUUUUGGAUUAUGAUUUCAUCUAAUUAUAUGAAAUUAUACAAAAACUGCAAUCGAUACGGUAACAGAUAUUUAGAUGAAAU_ ..((((.((((((...........)))))).)))).(((((((((((.((((......))))......(((..((((...))))..)))....))))))))))). (-17.05 = -17.50 + 0.45)

| Location | 1,026,992 – 1,027,097 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -17.84 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
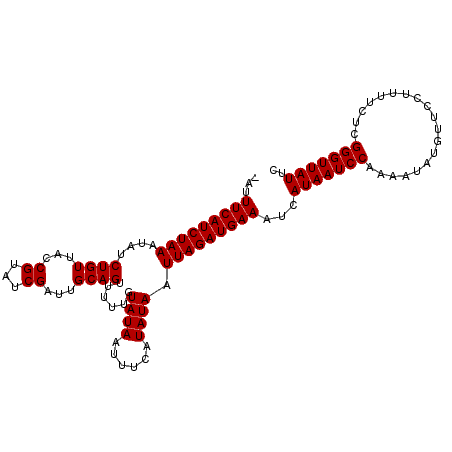
>2R_DroMel_CAF1 1026992 105 - 20766785 UAUUUCAUCUAAAUAUCUGUUACCGUAUCGAUUGCAGUUUUUGUAUAAUUUCAUAUAAUUAGAUGAAAUCAUAAUCCAAAAUAUGUUCCUUUUUUCGGGUUAUUC .(((((((((((....((((...((...))...))))...((((((......))))))))))))))))).(((((((((((..........)))).))))))).. ( -19.00) >DroSec_CAF1 29442 105 - 1 CAUUUCAUCUAAAUAUCUGUUACCGUAUCGAUUGCAGUUUUUGUAUAAUUUCAUAUAAUUAGAUGAAAUCAUAAUCCAAAAUAUGUUCCUUUUCUCGGGUUAUUC .(((((((((((....((((...((...))...))))...((((((......))))))))))))))))).(((((((...................))))))).. ( -17.81) >DroSim_CAF1 10932 97 - 1 --AUUCAUCUAAAUAUCUGUUACCGUAUCGAUUGCAG------UAUAAUUUCAUAUAAUUAGAUGAAAUCAUAAUCCAAAAUAUGUUCCUUUUCUCGGGUUAUUC --.(((((((((.(((.((.(((.(((.....))).)------))......)).))).)))))))))...(((((((...................))))))).. ( -16.71) >consensus _AUUUCAUCUAAAUAUCUGUUACCGUAUCGAUUGCAGUUUUUGUAUAAUUUCAUAUAAUUAGAUGAAAUCAUAAUCCAAAAUAUGUUCCUUUUCUCGGGUUAUUC ...(((((((((....((((...((...))...))))......((((......)))).)))))))))...(((((((...................))))))).. (-14.38 = -14.38 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:31 2006