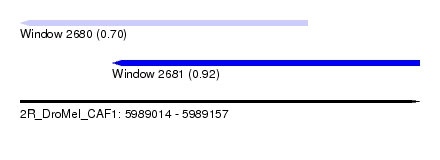
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,989,014 – 5,989,157 |
| Length | 143 |
| Max. P | 0.918076 |

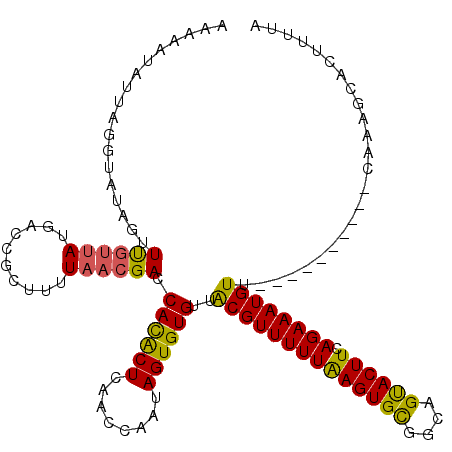
| Location | 5,989,014 – 5,989,117 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -16.60 |
| Energy contribution | -15.56 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5989014 103 - 20766785 CCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGUCGCAGUACUUCAGAAAUGUU----------CACAGCACUUUUAGUUCAAUAACUCAUGUGA-AUCCAUGUAAUAGUG ...((((.(((.((.(((((...(((((((((((((......))))).)))))))).----------....))))).)).)))........((((...-...))))....)))) ( -18.40) >DroSec_CAF1 7450 104 - 1 CCACACUCAACCAAUAGUGUGUUGCGUUUUUAAGUGCGGCCGUACUUAAGAAAUGUU----------CAAAGCACUUUUAGUUCAAUAACCCACGUGAUAUCCAUGUAAUAGCG .((((((........))))))..((.(((((((((((....))))))))))).....----------.........................(((((.....)))))....)). ( -22.50) >DroSim_CAF1 7942 103 - 1 CCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU----------CAAAGCACUUUUAGUUCAAUAACCCACGUGA-AUCCAUGUAAUAGCG ........(((.((.(((((...((((((((((((((....)))))).)))))))).----------....))))).)).))).........(((((.-...)))))....... ( -22.30) >DroEre_CAF1 7603 99 - 1 CCAUGCUCAACCAAAAGUGUGUUACGUUUUUAAGUGCAACAGCACUUCAGAAAUGUUUUCAUUGCUUUACGGUGCUUUCAGUGCAGAAACCUACU---------------AGCG ....(((..(((.((((((((..((((((((((((((....)))))).))))))))...)).))))))..)))((.......))...........---------------))). ( -23.60) >DroYak_CAF1 7628 113 - 1 CCAUACUCAACCAAAAGUGUGUUACGUUUUUGAGUGCGAUAGUACUUCAGAAAUGUUUUCAUUCCUACAAAUUACUUUCAGUGCAGAAAUCUACUUGC-AUCAAUGUAAAAGCG .((((((........))))))..((((((((((((((....)))).))))))))))...............((((.....((((((........))))-))....))))..... ( -24.50) >consensus CCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU__________CAAAGCACUUUUAGUUCAAUAACCCACGUGA_AUCCAUGUAAUAGCG .((((((........))))))..((((((((((((((....)))))).)))))))).......................................................... (-16.60 = -15.56 + -1.04)

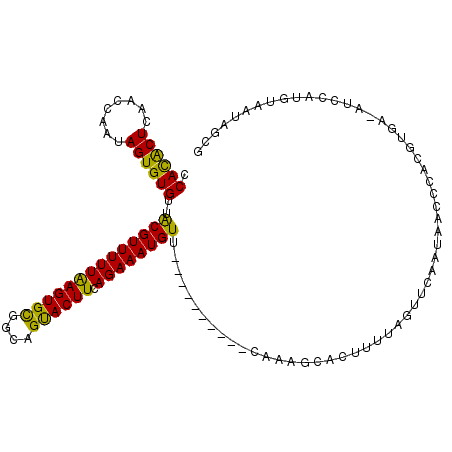
| Location | 5,989,047 – 5,989,157 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -17.48 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5989047 110 - 20766785 AAAAAUAUUAGGUAUACCUUGUUAUGACCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGUCGCAGUACUUCAGAAAUGUU----------CACAGCACUUUUA ....(((..(((....)))...)))(((.((((..((((..((((((........))))))...))))...)))))))((.((....((....))..----------.)).))....... ( -21.10) >DroSec_CAF1 7484 110 - 1 AAAAAUAUUAGGUAUAGUUCGUUAUGCCCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUGCGUUUUUAAGUGCGGCCGUACUUAAGAAAUGUU----------CAAAGCACUUUUA ..........(((((((....))))))).(((((.((((..((((((........)))))).....(((((((((((....))))))))))).))))----------.)))))....... ( -30.30) >DroSim_CAF1 7975 110 - 1 AAAAAUAUUAGGUAUAGUUCGUUAUGACCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU----------CAAAGCACUUUUA ..........(((..(((((((((..........))))))....)))..)))((.(((((...((((((((((((((....)))))).)))))))).----------....))))).)). ( -25.20) >DroEre_CAF1 7622 120 - 1 UAAAGUAUUAGGUAUAGUUUGUUAUGACCGUUUCUAACGACCAUGCUCAACCAAAAGUGUGUUACGUUUUUAAGUGCAACAGCACUUCAGAAAUGUUUUCAUUGCUUUACGGUGCUUUCA .((((((((.(((..(((((((((.((.....))))))))....)))..))).((((((((..((((((((((((((....)))))).))))))))...)).))))))..)))))))).. ( -29.20) >DroYak_CAF1 7661 112 - 1 --------UAAACAGCAUUUUAAAUGACCGUUUCUAACGACCAUACUCAACCAAAAGUGUGUUACGUUUUUGAGUGCGAUAGUACUUCAGAAAUGUUUUCAUUCCUACAAAUUACUUUCA --------..............(((((.(((.....)))..((((((........))))))..((((((((((((((....)))).))))))))))..)))))................. ( -20.20) >consensus AAAAAUAUUAGGUAUAGUUUGUUAUGACCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU__________CAAAGCACUUUUA ..................((((((..........)))))).((((((........))))))..((((((((((((((....)))))).))))))))........................ (-17.48 = -16.80 + -0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:32 2006