| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,963,951 – 5,964,052 |
| Length | 101 |
| Max. P | 0.859204 |

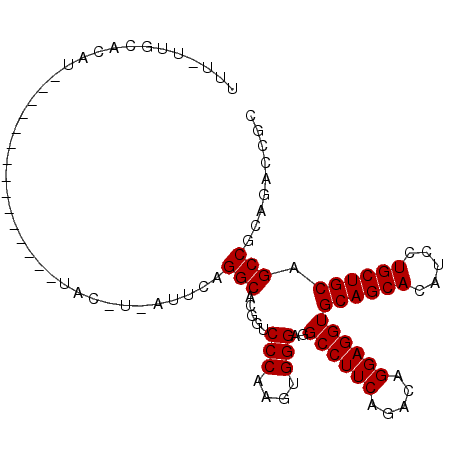
| Location | 5,963,951 – 5,964,052 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5963951 101 + 20766785 GCGGUCUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGGCCAUGCCUGAAUGAGGUACAA---AAAAAAAUAAUGUGCAA-AAA (((((....((((.((((((((.(((...))).))))).)))..)))).(((....)))))).(((((.....)))))...---.............))..-... ( -32.90) >DroPse_CAF1 3102 82 + 1 GCCGUUUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGCCUGAAGGCCUCCCACUUGGGUCCGUGCCUGCG----------------------GUGGGCAA-GAG ((((....))))(((((((.....))))))).(((.((((((.(((((.(((....)))...).))))...----------------------.)))))).-))) ( -37.70) >DroSim_CAF1 6632 102 + 1 GCGGUCUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCAUGCCUGAAUAAGGUACAACAAAAAAAAA--AUGUGCAA-AAA ((((((...((((.((((((((.(((...))).))))).)))..))))..((....)))))).(((((.....))))).............--....))..-... ( -33.80) >DroEre_CAF1 6675 82 + 1 GCGGUCUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCGUGCCUGAAUAAAGUAG-----------------------AAU ....(((((((((.((((((((.(((...))).))))).)))..))))((((....))))...............))))-----------------------).. ( -30.10) >DroYak_CAF1 6844 95 + 1 GCGGUCUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCAUGCCUGAAUGAAGUAA--------AAAA--AAGUGCAAAAAA ..((((...((((.((((((((.(((...))).))))).)))..))))..((....)))))).(((((...........--------....--.)).)))..... ( -28.69) >DroPer_CAF1 3107 82 + 1 GCCGUUUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGUCCGUGCCUGCG----------------------GUGGGCAA-GAG ((((....))))(((((((.....))))))).(((.((((((.(((((.(((....)))...).))))...----------------------.)))))).-))) ( -35.00) >consensus GCGGUCUGCGGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCAUGCCUGAAU_A_GUA_______________AUGUGCAA_AAA (((((.....))))).((((((.(((...))).))))))....((((.((((....))))....))))..................................... (-24.84 = -25.28 + 0.45)

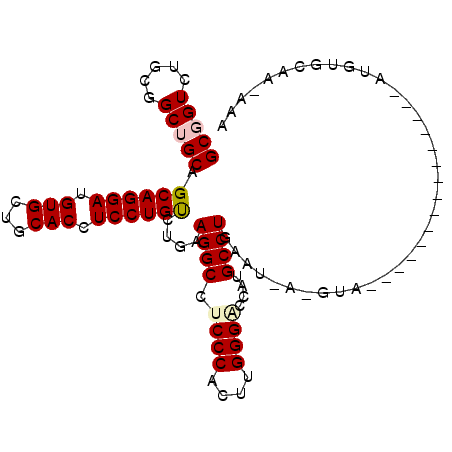
| Location | 5,963,951 – 5,964,052 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -26.27 |
| Energy contribution | -26.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5963951 101 - 20766785 UUU-UUGCACAUUAUUUUUUU---UUGUACCUCAUUCAGGCAUGGCCCCAAGUGGGAGGCCUUCAGACAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAGACCGC ..(-((((...........((---((((.(((.....)))...((((((.....)).)))).....))))))(((((((((.....))))))).))))))).... ( -32.20) >DroPse_CAF1 3102 82 - 1 CUC-UUGCCCAC----------------------CGCAGGCACGGACCCAAGUGGGAGGCCUUCAGGCAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAAACGGC (((-(((((...----------------------.(.((((.(...(((....))).))))).).)))))))).(((((((.....)))))))((((....)))) ( -37.50) >DroSim_CAF1 6632 102 - 1 UUU-UUGCACAU--UUUUUUUUUGUUGUACCUUAUUCAGGCAUGGUCCCAAGUGGGAGGCCUUCAGACAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAGACCGC ...-..((....--........(((((((((((..((((((....((((....)))).))))...))...))))))))))).......((((....))))...)) ( -32.70) >DroEre_CAF1 6675 82 - 1 AUU-----------------------CUACUUUAUUCAGGCACGGUCCCAAGUGGGAGGCCUUCAGACAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAGACCGC ...-----------------------................(((((....((((...((((((.....))))))((((((.....))))))..)))).))))). ( -28.60) >DroYak_CAF1 6844 95 - 1 UUUUUUGCACUU--UUUU--------UUACUUCAUUCAGGCAUGGUCCCAAGUGGGAGGCCUUCAGACAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAGACCGC ...(((((....--....--------..(((((..((((((....((((....)))).))))...))...)))))((((((.....))))))....))))).... ( -30.00) >DroPer_CAF1 3107 82 - 1 CUC-UUGCCCAC----------------------CGCAGGCACGGACCCAAGUGGGAGGCCUUCAGACAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAAACGGC .((-(((((...----------------------....)))).)))(((....)))..((((((.....))))))((((((.....)))))).((((....)))) ( -31.10) >consensus UUU_UUGCACAU_______________UAC_U_AUUCAGGCACGGUCCCAAGUGGGAGGCCUUCAGACAGGAGGUGCAGCACAUCCUGCUGCAGCCGCAGACCGC ......................................(((.....(((....)))..((((((.....))))))((((((.....)))))).)))......... (-26.27 = -26.27 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:27 2006