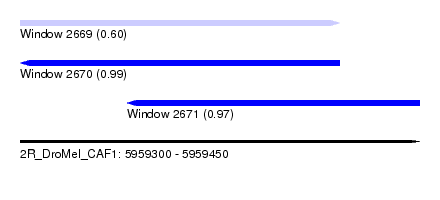
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,959,300 – 5,959,450 |
| Length | 150 |
| Max. P | 0.991771 |

| Location | 5,959,300 – 5,959,420 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.66 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5959300 120 + 20766785 UAGUGGGAAACGAAAAUGGGCAAAUGAAAUUAGCAUUUAUCAAAUUAUUCCAUUUUCCCAAGGACAGCUUGGCUAAGCAAUUUUCCAGCCGAGCUGCCAGUUAUCUUGAUUAUCUCGUUU .......((((((...((((.(((((.(((................))).))))).)))).((.((((((((((............))))))))))))................)))))) ( -31.99) >DroSec_CAF1 1922 120 + 1 UAGUGGGAAGCGAAAAUGGGCAAAUGAAAUUAGCAUUUAUCAAAUUAUGCCAUUUUCCCAAGGACAGCUAGGCUAAGCAAUUUUCCAGCCGAGCUGCCAGCUAUCUUGAUUAUCUCGUUU (((((((((((.....((((.(((((......((((..........))))))))).)))).((.(((((.((((............)))).))))))).))).)))).))))........ ( -31.00) >DroSim_CAF1 2026 120 + 1 UAGUGGCAAGCGAAAAUGGGCAAAUGAAAUUACCAUUUAUCAAAUUAUGCCAUUUUCCCAAGGACAGCUAGGCUAAGCAAUUUUCCAGCCGAGCUGCCAGCUAUCUUGAUUAUCUCGUUU ..(((((..(.((((((((.((((((.......))))..........)))))))))).)..((.(((((.((((............)))).))))))).)))))................ ( -29.40) >DroEre_CAF1 2066 120 + 1 UAGUAGGCAGCGAAAAUAGGCAAAUGAAAUUACCAUUUAUCAAAUUAUGCCAUUUUCCCAAGGACAGCUAGGCUAAGCAAUUUUCCAACCGAGCUGCCUGCUAUCUUGAUUAUCUCGUUU (((((((((((((((((.((((((((.......))))..........))))))))))....(((..(((......))).....)))......)))))))))))................. ( -32.70) >DroYak_CAF1 1960 119 + 1 UAGUGGGCAGCAGAAAUAGGCAAAUGAAAUAACCAUUUAUCAAAUUGUGCUGUUUUC-CAAGGACAGCUAGACUAAGCAAUUUUCCAGCCGAGCUGCCAGCUAUCUUGAUUAUCUCGUUU ((((.(((((((....).((((((((.......)))))...((((((((((((((..-...)))))))........)))))))....)))..)))))).))))................. ( -31.20) >consensus UAGUGGGAAGCGAAAAUGGGCAAAUGAAAUUACCAUUUAUCAAAUUAUGCCAUUUUCCCAAGGACAGCUAGGCUAAGCAAUUUUCCAGCCGAGCUGCCAGCUAUCUUGAUUAUCUCGUUU ....((..(((((((((.((((((((.......))))..........))))))))))....((.(((((.((((............)))).))))))).)))..)).............. (-22.82 = -22.66 + -0.16)

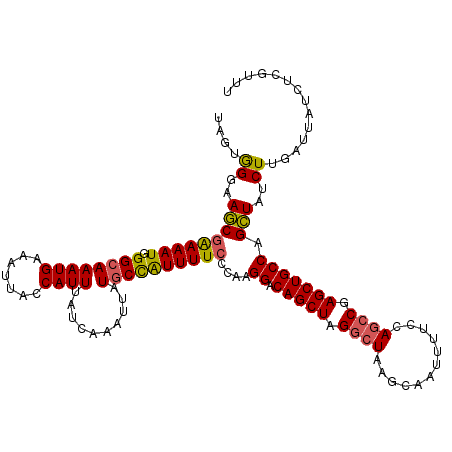
| Location | 5,959,300 – 5,959,420 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.24 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5959300 120 - 20766785 AAACGAGAUAAUCAAGAUAACUGGCAGCUCGGCUGGAAAAUUGCUUAGCCAAGCUGUCCUUGGGAAAAUGGAAUAAUUUGAUAAAUGCUAAUUUCAUUUGCCCAUUUUCGUUUCCCACUA ((((((((..............(((((((.((((((........)))))).)))))))..((((.(((((((((................))))))))).)))).))))))))....... ( -35.79) >DroSec_CAF1 1922 120 - 1 AAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUGAUAAAUGCUAAUUUCAUUUGCCCAUUUUCGCUUCCCACUA ...(((((...(((((((((((((..(((.(((.........))).))))))))))).)))))((((.((((((..........)))))).))))..........))))).......... ( -32.20) >DroSim_CAF1 2026 120 - 1 AAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUGAUAAAUGGUAAUUUCAUUUGCCCAUUUUCGCUUGCCACUA ...((((....(((((((((((((..(((.(((.........))).))))))))))).)))))((((((((((.....)).(((((((.....))))))).)))))))).))))...... ( -35.00) >DroEre_CAF1 2066 120 - 1 AAACGAGAUAAUCAAGAUAGCAGGCAGCUCGGUUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUGAUAAAUGGUAAUUUCAUUUGCCUAUUUUCGCUGCCUACUA ..............((.((((.(((((((.((((((........)))))).))))))).....((((((((((.....(((............)))..)))).)))))))))).)).... ( -32.00) >DroYak_CAF1 1960 119 - 1 AAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGUCUAGCUGUCCUUG-GAAAACAGCACAAUUUGAUAAAUGGUUAUUUCAUUUGCCUAUUUCUGCUGCCCACUA ...........(((((((((((((..(((.(((.........))).))))))))))).))))-)....(((((.((...(.(((((((.....))))))).)...)).)))))....... ( -30.00) >consensus AAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUGAUAAAUGGUAAUUUCAUUUGCCCAUUUUCGCUUCCCACUA ...............(..(((.(((((((.((((((........)))))).))))))).....((((((((((.....)).(((((((.....))))))).)))))))))))..)..... (-27.44 = -27.24 + -0.20)

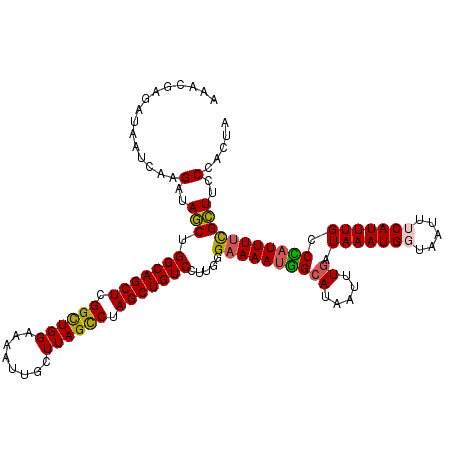
| Location | 5,959,340 – 5,959,450 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5959340 110 - 20766785 UUGAGUGUCUACUGAAGGGGGUU-UUGUGAUAAACGAGAUAAUCAAGAUAACUGGCAGCUCGGCUGGAAAAUUGCUUAGCCAAGCUGUCCUUGGGAAAAUGGAAUAAUUUG ..((((.((((((.((((..(((-((((.....)))))))..............((((((.((((((........)))))).)))))))))).))....))))...)))). ( -29.70) >DroSec_CAF1 1962 110 - 1 UUGAGUGUCUGCUGAAGGGGGUU-UUGUGAUAAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUG ....(((((..((.((((..(((-((((.....)))))))..............((((((.((((((........)))))).)))))))))).)).....)))))...... ( -32.60) >DroSim_CAF1 2066 110 - 1 UUGAGUGUCUGCUGAAGGGGGUU-UUGUGAUAAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUG ....(((((..((.((((..(((-((((.....)))))))..............((((((.((((((........)))))).)))))))))).)).....)))))...... ( -32.60) >DroEre_CAF1 2106 110 - 1 UUGAGUGUCUGCUGAAGGGGGUU-UUGUGAUAAACGAGAUAAUCAAGAUAGCAGGCAGCUCGGUUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUG ....((((((((((..((..(((-((((.....)))))))..))....)))))(((((((.((((((........)))))).)))))))...........)))))...... ( -31.30) >DroYak_CAF1 2000 110 - 1 UUGAGUGUCUGCUGAAGGGGGUUUUUGUGAUAAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGUCUAGCUGUCCUUG-GAAAACAGCACAAUUUG ..((((...(((((........((((((.....))))))...(((((((((((((..(((.(((.........))).))))))))))).))))-)....)))))..)))). ( -30.30) >consensus UUGAGUGUCUGCUGAAGGGGGUU_UUGUGAUAAACGAGAUAAUCAAGAUAGCUGGCAGCUCGGCUGGAAAAUUGCUUAGCCUAGCUGUCCUUGGGAAAAUGGCAUAAUUUG ....(((((..((.((((.((((.((((.....))))...))))..........((((((.((((((........)))))).)))))))))).)).....)))))...... (-28.06 = -28.18 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:23 2006