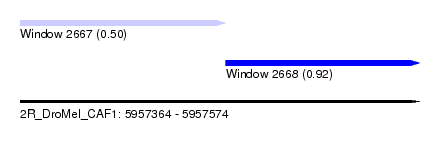
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,957,364 – 5,957,574 |
| Length | 210 |
| Max. P | 0.917496 |

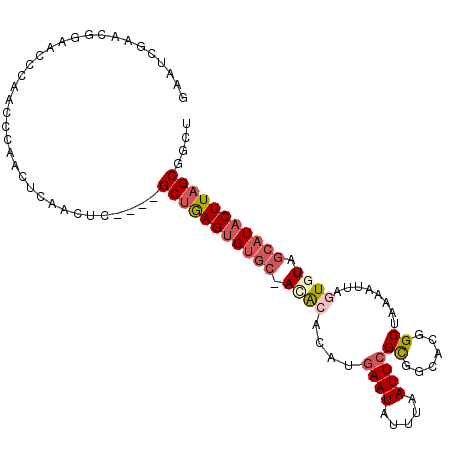
| Location | 5,957,364 – 5,957,472 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5957364 108 + 20766785 AGUCGCCUCAUCGAGCGCAAUCUCAAGGAGGACGGCAUACAGGCGGCAAAGGACAUCAAACUCCUGCUGCUGGGUGAGUUGUCCCCGGGAACG--UAAUC-GAAACCAAAU ..........((((..((..((((..((.(((((((.(((.(((((((..(((........))))))))))..))).)))))))))))))..)--)..))-))........ ( -37.10) >DroSim_CAF1 46 108 + 1 AGUCGCCUCAUCGAGCGCAAUCUCAAGGAGGACGGCAUACAGGCGGCAAAGGACAUCAAACUCCUGCUGCUGGGUGAGUUUUCCCCGGGAACG--CAAUC-GAAACAAACU ..........((((..((..((((..(((((((..(((...(((((((..(((........))))))))))..))).)))))))..))))..)--)..))-))........ ( -35.60) >DroEre_CAF1 46 110 + 1 AGUCGCCUCAUCGAGCGCAAUCUCAAGGAGGACGGCAUACAGGCGGCAAAGGACAUCAAACUUCUGCUUCUGGGUGAGUUUUCCCCGGGAACGAACAAUC-GAAACAAUUC ....(((((...))).))..((((..(((((((..((..((((.((((.(((........))).))))))))..)).)))))))..)))).(((....))-)......... ( -28.90) >DroWil_CAF1 5925 99 + 1 AGUCGUCUAAUUGAACGUAAUCUUAAAGAAGAUGGCAUACAGGCGGCAAAGGACAUUAAACUUUUGCUACUGGGUGAGUGUGU---------GAAUAAGCCGUGAAAA--- .(((((((..((((........))))...)))))))...(((..((((((((........)))))))).)))(((...(((..---------..))).))).......--- ( -23.60) >DroYak_CAF1 46 97 + 1 AGUCGCCUCAUCGAGCGCAAUCUCAAGGAGGACGGCAUACAGGCGGCAAAGGACAUCAAACUUCUGCUGCUGGGUGAG-----------AACG--CAAUC-GAAACAAAUU .((((((((...(((......)))...)))).)))).(((.(((((((.(((........))).)))))))..)))..-----------....--.....-.......... ( -28.00) >DroAna_CAF1 46 107 + 1 AGUCGCCUCAUCGAGCGCAAUCUCAAGGAGGACGGCAUACAGGCGGCAAAGGACAUCAAGCUCCUGCUGUUGGGUGAGUUUGG-AAGAGGACGAAGAGGCCCAUAAAA--- .((((((((...(((......)))...)))).)))).....(((((((..(((........))))))))))((((...((((.-.......))))...))))......--- ( -31.70) >consensus AGUCGCCUCAUCGAGCGCAAUCUCAAGGAGGACGGCAUACAGGCGGCAAAGGACAUCAAACUCCUGCUGCUGGGUGAGUUUUC_CCGGGAACG__CAAUC_GAAACAAA_U .((((((((...(((......)))...)))).)))).(((.(((((((..(((........))))))))))..)))................................... (-21.58 = -21.47 + -0.11)

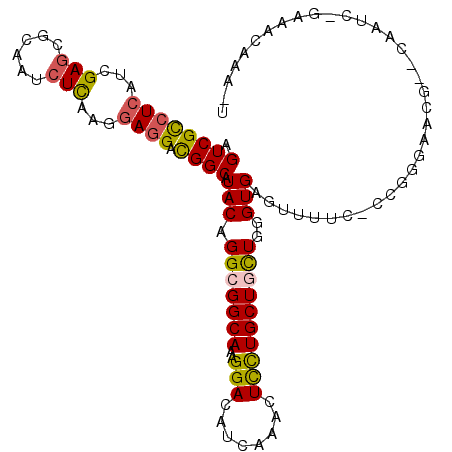
| Location | 5,957,472 – 5,957,574 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.90 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5957472 102 + 20766785 GAAUCGAACGGAAACCAACCCAACUCAACUC----GCUGAGUGUGCCACACACAUGAAUAUUUGAUUCGCGGCACGGCUAAAAUUAGUGUAGCAUACUUAGCGGCU .........((........))........((----(((((((((((.((((....((((.....))))((......))........)))).))))))))))))).. ( -30.10) >DroSec_CAF1 154 101 + 1 GAAUCGAACGGAACCCAACCCAACUCAACUC----GCUGAGUGUGC-ACAGACAUGAAUAUUUGAUUCGCGGCACGGCUAAAAUUAGUGUAGCAUACUUAGCGGCU .........((........))........((----(((((((((((-.....(.(((((.....))))).)((((...........)))).))))))))))))).. ( -27.00) >DroSim_CAF1 154 102 + 1 GAAUCGAACGGAACCCAACCCAACUCAACUC----GCUGAGUGUGCCACACACAUGAAUAUUUGAUUCGCGGCACGGCUAAAAUUAGUGUAGCAUACUUAGCGGCU .........((........))........((----(((((((((((.((((....((((.....))))((......))........)))).))))))))))))).. ( -30.10) >DroEre_CAF1 156 91 + 1 GAAUCGAACGUAACCC----------AACUC----GCUGAGUGUGC-AUACACAUGAAUAUUUAAUUCGCUGCUUGGCUAAAAUUAGUGUAGCAUACUUAGCGGCU ................----------...((----(((((((((((-.(((((..((((.....))))(((....)))........)))))))))))))))))).. ( -30.50) >DroYak_CAF1 143 91 + 1 GAAUCGAACGAAACCC----------AACUC----GCUGAGUGUGC-AUACACAUGAAUAUUUAAUUCGCUGCUCGGCUAAAUUUAGUCUAGCAUACUUAGCAACU ................----------.....----(((((((((((-.....(((((((.....))))).))...(((((....)))))..))))))))))).... ( -24.20) >DroAna_CAF1 153 104 + 1 -ACUCGUUGGUAACUCCGUUCUUGUCGACCUGCGGGCCAAGGGCCA-AGGGCCAUAAAUAUUUAAUUCGUGCCUCUGCUAAAAUUAGUGUAGCAUACUUAGCGGCU -...(.(((((....((((............))))))))).)(((.-((((.((..(((.....)))..)).))))((((.....(((((...)))))))))))). ( -23.60) >consensus GAAUCGAACGGAACCCAACCCAACUCAACUC____GCUGAGUGUGC_ACACACAUGAAUAUUUAAUUCGCGGCACGGCUAAAAUUAGUGUAGCAUACUUAGCGGCU ...................................(((((((((((.((((....((((.....))))((......))........)))).))))))))))).... (-18.73 = -19.90 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:20 2006