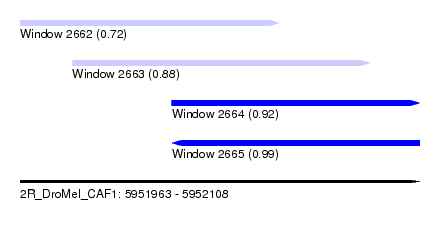
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,951,963 – 5,952,108 |
| Length | 145 |
| Max. P | 0.989510 |

| Location | 5,951,963 – 5,952,057 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.08 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.72 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5951963 94 + 20766785 GCGAAACCGAAGU-A----------------AUUUGGGUCAACUUUUGGGUGUUGCAUGUGGCAUGGAUAUCCGAAAAUAAAAUGGACUCAUG-CACCACUGUGCAUGCAAC ((((..(((((..-.----------------.)))))(((...(((((((((((.((((...)))))))))))))))........)))))(((-(((....))))))))... ( -27.50) >DroPse_CAF1 11099 109 + 1 GCUAAACCGAAACCACAACAAAAGAGUACCCAUUUGGGGCAGCUCUUUUG-CUUGCAUGUCGAAUUACUUUCACGCGCUGAAGGGAUGUCACGGAACCACUGUGCAGGCA-- (((....(((.....((((((((((((.(((....)))...)))))))))-.)))....))).....((((((.....))))))..((.(((((.....)))))))))).-- ( -32.30) >DroSec_CAF1 6932 94 + 1 GCGAAACCGAAGU-A----------------AUUUGGGUCAACUUUUGGGUGUUGCAUGUGGCAUGGAUAUCCGAAAAUAAAAUGAAAUCAUG-CACCACUGUGCAUGCAAC ((((..(((((..-.----------------.)))))......(((((((((((.((((...)))))))))))))))...........))(((-(((....))))))))... ( -27.00) >DroSim_CAF1 7396 94 + 1 GCGAAACCGAAGU-A----------------AUUUGGGUCAACUUUUGGGUGUUGCAUGUGGCAUGGAUAUCCGAAAAUAAAAUGGACGCAUG-CACCACUGUGCAUGCAAC ......(((((..-.----------------.)))))(((...(((((((((((.((((...)))))))))))))))........)))(((((-(((....))))))))... ( -32.60) >DroEre_CAF1 6689 94 + 1 GCGAAACCGCAGU-A----------------AUUUGGGUCAACUUUUGGCUGUUGCAUGUGGAAAGUAUAUCCCAAAAUAAAAUGAACUCAUG-CACCACUGUGCAUGCAAC (((....)))...-.----------------.((((((...((((((.((........)).))))))....))))))............((((-(((....))))))).... ( -27.20) >DroPer_CAF1 10954 109 + 1 GCUAAACCGAAACCACAACAAAAGAGUACCCAUUUGGGGCAGCUCUUCUG-CUUGCAUGUCGAAUUACUUUCACGCGUUGAAGGGAUGUCACGGAACCACUGUGCAGGCA-- .....................((((((.(((....)))...))))))..(-(((((((((.((....((((((.....))))))....))))(....)...)))))))).-- ( -28.80) >consensus GCGAAACCGAAGU_A________________AUUUGGGUCAACUUUUGGGUGUUGCAUGUGGAAUGGAUAUCCCAAAAUAAAAUGAACUCAUG_CACCACUGUGCAUGCAAC (((...(((((.....................)))))................(((((((((...(......).........(((....)))....)))).))))))))... (-11.21 = -10.72 + -0.50)


| Location | 5,951,982 – 5,952,090 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.66 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -12.53 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5951982 108 + 20766785 GGUCAACUUUUGGGUGUUGCAUGUGGCAUGG--A--UAUCCGAAAAUAAAAUGGACUCAUG-CACCACUGUGCAUGCAACAUCUUAGACAAUGG-------CAGCAGCAACAGUUGCAUC .((((..(((.(((((((((((((.((((((--.--..((((.........)))).)))))-)((....))))))))))))))).)))...)))-------).(((((....)))))... ( -36.20) >DroSec_CAF1 6951 108 + 1 GGUCAACUUUUGGGUGUUGCAUGUGGCAUGG--A--UAUCCGAAAAUAAAAUGAAAUCAUG-CACCACUGUGCAUGCAACAUCUUAGACAAUGG-------CAGCAGCAACAGUUGCAUC .((((..(((.(((((((((((((.(((((.--(--(.((............)).))))))-)((....))))))))))))))).)))...)))-------).(((((....)))))... ( -32.80) >DroSim_CAF1 7415 108 + 1 GGUCAACUUUUGGGUGUUGCAUGUGGCAUGG--A--UAUCCGAAAAUAAAAUGGACGCAUG-CACCACUGUGCAUGCAACAUCUUAGACAAUGG-------CAGCAGCAACAGUUGCAUC .((((..(((((((((((.((((...)))))--)--))))))))).(((.(((...(((((-(((....))))))))..))).))).....)))-------).(((((....)))))... ( -37.00) >DroEre_CAF1 6708 108 + 1 GGUCAACUUUUGGCUGUUGCAUGUGGAAAGU--A--UAUCCCAAAAUAAAAUGAACUCAUG-CACCACUGUGCAUGCAACAUCUUAGACAAUGG-------CAGCAGCAACAGUUGCAUC ((((((((....((((((((.((.(((....--.--..)))))...(((.(((....((((-(((....)))))))...))).))).......)-------)))))))...))))).))) ( -31.90) >DroAna_CAF1 6731 103 + 1 GGUCAUGUUUUCGG-GUUGCAUGUCGAGUUUCGAAGCCUCCAAAAAUAAAG-----UCUCG-UACCAAUGUGC----AACA------ACAAUGGAAUGCCACAGCAGCAACAGUUGCAUC ((.(((........-(((((((((.(.((..(((..(.............)-----..)))-.))).))))))----))).------........)))))...(((((....)))))... ( -24.15) >DroPer_CAF1 10990 93 + 1 GGGCAGCUCUUCUG-CUUGCAUGUCGAAUUA--C--UUUCACGCGUUGAAGGGAUGUCACGGAACCACUGUGCAGGCA---------------G-------CAGCAGCAACAGUUGCAUC ..((((((.(((((-((.((.((((......--(--(((((.....))))))..((.(((((.....)))))))))))---------------)-------)))))).)).))))))... ( -33.40) >consensus GGUCAACUUUUGGGUGUUGCAUGUGGAAUGG__A__UAUCCGAAAAUAAAAUGAACUCAUG_CACCACUGUGCAUGCAACAUCUUAGACAAUGG_______CAGCAGCAACAGUUGCAUC ...............(((((((((..........................(((....)))...((....))))))))))).......................(((((....)))))... (-12.53 = -13.75 + 1.22)
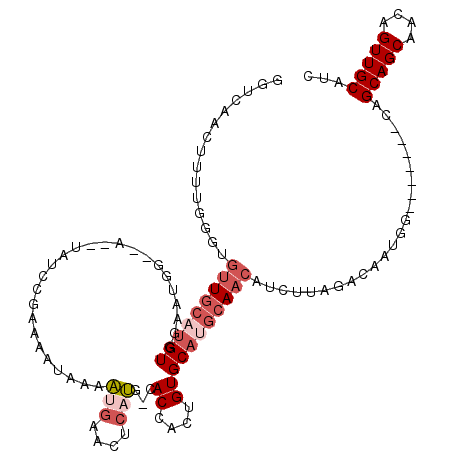

| Location | 5,952,018 – 5,952,108 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -19.08 |
| Energy contribution | -20.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
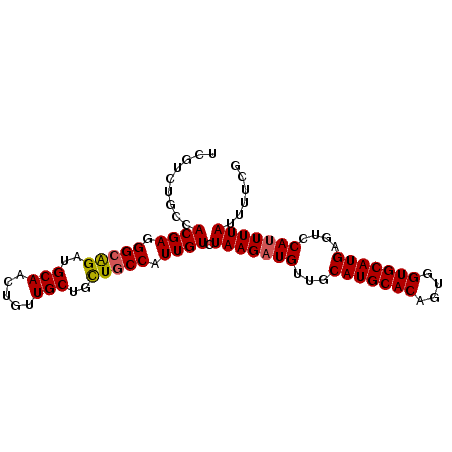
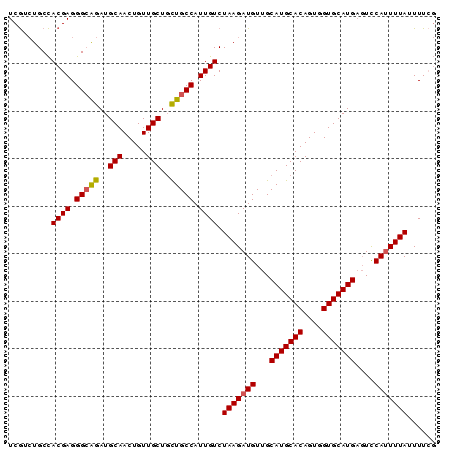
>2R_DroMel_CAF1 5952018 90 + 20766785 CGAAAAUAAAAUGGACUCAUGCACCACUGUGCAUGCAACAUCUUAGACAAUGGCAGCAGCAACAGUUGCAUCCGCCCUCGUGGCAGGCGA ......(((.(((....(((((((....)))))))...))).))).......((.(((((....)))))....(((.....)))..)).. ( -23.40) >DroSec_CAF1 6987 90 + 1 CGAAAAUAAAAUGAAAUCAUGCACCACUGUGCAUGCAACAUCUUAGACAAUGGCAGCAGCAACAGUUGCAUCUGCCCUCGUGGCAGACGA ((....(((.(((....(((((((....)))))))...))).)))..........(((((....))))).((((((.....)))))))). ( -25.10) >DroSim_CAF1 7451 90 + 1 CGAAAAUAAAAUGGACGCAUGCACCACUGUGCAUGCAACAUCUUAGACAAUGGCAGCAGCAACAGUUGCAUCUGCCCUCGUGGCAGACGA ((....(((.(((...((((((((....))))))))..))).)))..........(((((....))))).((((((.....)))))))). ( -30.00) >DroEre_CAF1 6744 90 + 1 CCAAAAUAAAAUGAACUCAUGCACCACUGUGCAUGCAACAUCUUAGACAAUGGCAGCAGCAACAGUUGCAUCUUCCCUCGUGGCAGCCGA ......(((.(((....(((((((....)))))))...))).))).....((((.(((((....)))))......((....))..)))). ( -22.60) >DroYak_CAF1 6941 88 + 1 --AAAAUAAAGUGGACUCAUGCACUACUGUGCAUGCAACAUCUUAGACAAUGGCAAGAGCAACAGUUGCAUCUGCCCUCGUAGCAUUUGA --......(((((.((.(((((((....)))))))................((((.(((((.....))).))))))...))..))))).. ( -21.70) >consensus CGAAAAUAAAAUGGACUCAUGCACCACUGUGCAUGCAACAUCUUAGACAAUGGCAGCAGCAACAGUUGCAUCUGCCCUCGUGGCAGACGA ......(((.(((....(((((((....)))))))...))).))).......(((((.......))))).((((((.....))))))... (-19.08 = -20.08 + 1.00)



| Location | 5,952,018 – 5,952,108 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5952018 90 - 20766785 UCGCCUGCCACGAGGGCGGAUGCAACUGUUGCUGCUGCCAUUGUCUAAGAUGUUGCAUGCACAGUGGUGCAUGAGUCCAUUUUAUUUUCG .........((((.(((((..(((.....)))..))))).)))).(((((((...(((((((....)))))))....)))))))...... ( -27.40) >DroSec_CAF1 6987 90 - 1 UCGUCUGCCACGAGGGCAGAUGCAACUGUUGCUGCUGCCAUUGUCUAAGAUGUUGCAUGCACAGUGGUGCAUGAUUUCAUUUUAUUUUCG .(((((...((((.(((((..(((.....)))..))))).))))...)))))...(((((((....)))))))................. ( -29.30) >DroSim_CAF1 7451 90 - 1 UCGUCUGCCACGAGGGCAGAUGCAACUGUUGCUGCUGCCAUUGUCUAAGAUGUUGCAUGCACAGUGGUGCAUGCGUCCAUUUUAUUUUCG .(((((...((((.(((((..(((.....)))..))))).))))...)))))..((((((((....))))))))................ ( -33.10) >DroEre_CAF1 6744 90 - 1 UCGGCUGCCACGAGGGAAGAUGCAACUGUUGCUGCUGCCAUUGUCUAAGAUGUUGCAUGCACAGUGGUGCAUGAGUUCAUUUUAUUUUGG ..(((.((..(((.((.........)).)))..)).)))......(((((((...(((((((....)))))))....)))))))...... ( -25.80) >DroYak_CAF1 6941 88 - 1 UCAAAUGCUACGAGGGCAGAUGCAACUGUUGCUCUUGCCAUUGUCUAAGAUGUUGCAUGCACAGUAGUGCAUGAGUCCACUUUAUUUU-- .........((((.(((((..(((.....)))..))))).))))....(((....(((((((....))))))).)))...........-- ( -24.10) >consensus UCGUCUGCCACGAGGGCAGAUGCAACUGUUGCUGCUGCCAUUGUCUAAGAUGUUGCAUGCACAGUGGUGCAUGAGUCCAUUUUAUUUUCG .........((((.(((((..(((.....)))..))))).)))).(((((((...(((((((....)))))))....)))))))...... (-26.14 = -26.22 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:17 2006