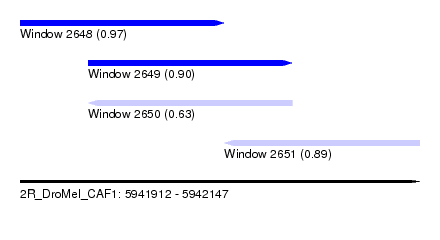
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,941,912 – 5,942,147 |
| Length | 235 |
| Max. P | 0.974568 |

| Location | 5,941,912 – 5,942,032 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -28.61 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5941912 120 + 20766785 UGUAGAAUAAUCAAAACUGACUUGCUAGCAAUGUUAAAAAGUGCAGAAAGCAUUGGCUAAUAUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUU ..............(((((....(((((((.(((...(((((((((((((((.(((((((......))))))).)).)))))))))))))...)))))).......))))..)))))... ( -32.71) >DroSec_CAF1 83994 120 + 1 UGUAGAAUAAUCAAAGUUGACUUGCUAGGAAUGUUAAUAAGUGCAGAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUU .....((((((...(((((..((((.....((((....((((((((((((((.(((((((......))))))).)).))))))))))))....)))))))).....)))))...)))))) ( -31.30) >DroSim_CAF1 81841 120 + 1 UGUAGAAUUAUCAAAAUUGACUUGCUAGCAAUGUUAAAAAGUGCAGAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUU .....((((((((((((((...(((((((...)))).(((((((((((((((.(((((((......))))))).)).)))))))))))))...)))..)))))))).....))))))... ( -32.00) >DroYak_CAF1 86747 120 + 1 UGUAGAAUAAUCAAAACUAACUUGCUAGCAAUAUUUAAAAGUGCAAAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUU .................(((((.((.(((........(((((((((((((((.(((((((......))))))).)).)))))))))))))..((((........)))))))))))))).. ( -30.10) >consensus UGUAGAAUAAUCAAAACUGACUUGCUAGCAAUGUUAAAAAGUGCAGAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUU ..................((((.(((((((.(((...(((((((((((((((.(((((((......))))))).)).)))))))))))))...)))))).......))))...))))... (-28.61 = -28.98 + 0.37)


| Location | 5,941,952 – 5,942,072 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -40.66 |
| Energy contribution | -40.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.02 |
| Mean z-score | -4.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5941952 120 + 20766785 GUGCAGAAAGCAUUGGCUAAUAUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUUGCUGUUGUAUUUUUUUGUGCUGCAGCUGCGCGUCGCAACU ((((((((((((.(((((((......))))))).)).))))))))))....(((.((((......(((((((((((...(((....))).........)))))))))))))))))).... ( -40.50) >DroSec_CAF1 84034 120 + 1 GUGCAGAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUUGCUGUUGUAUUUUUUUGUGCUGCAGCUGCGCGUCGCAACU ((((((((((((.(((((((......))))))).)).))))))))))....(((.((((......(((((((((((...(((....))).........)))))))))))))))))).... ( -40.50) >DroSim_CAF1 81881 120 + 1 GUGCAGAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUUGCUGUUGUAUUUUUUUGUGCUGCAGCUGCGCGUCGCAACU ((((((((((((.(((((((......))))))).)).))))))))))....(((.((((......(((((((((((...(((....))).........)))))))))))))))))).... ( -40.50) >DroYak_CAF1 86787 119 + 1 GUGCAAAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUUGCUGUUGUAUUU-UUUGUGCUGCAGCUGCGCGUCGCAACU ((((((((((((.(((((((......))))))).)).))))))))))....(((.((((......(((((((((((...(((....)))...-.....)))))))))))))))))).... ( -41.00) >consensus GUGCAGAAAGCAUUGGCUAAUUUCGUUUGGCUAUUGAUUUUUUGCAUUUUCGUGCAUGCAAUUUUGUAGCUGUAGUUGUUGCUGUUGUAUUUUUUUGUGCUGCAGCUGCGCGUCGCAACU ((((((((((((.(((((((......))))))).)).))))))))))....(((.((((......(((((((((((...(((....))).........)))))))))))))))))).... (-40.66 = -40.48 + -0.19)


| Location | 5,941,952 – 5,942,072 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5941952 120 - 20766785 AGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGCAACAACUACAGCUACAAAAUUGCAUGCACGAAAAUGCAAAAAAUCAAUAGCCAAACGAUAUUAGCCAAUGCUUUCUGCAC (((((((...)))))))((((.............................((((.....((((((........))))))........))))...........(((....)))..)))).. ( -20.82) >DroSec_CAF1 84034 120 - 1 AGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGCAACAACUACAGCUACAAAAUUGCAUGCACGAAAAUGCAAAAAAUCAAUAGCCAAACGAAAUUAGCCAAUGCUUUCUGCAC (((((((...)))))))((((.............................((((.....((((((........))))))........))))...........(((....)))..)))).. ( -20.82) >DroSim_CAF1 81881 120 - 1 AGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGCAACAACUACAGCUACAAAAUUGCAUGCACGAAAAUGCAAAAAAUCAAUAGCCAAACGAAAUUAGCCAAUGCUUUCUGCAC (((((((...)))))))((((.............................((((.....((((((........))))))........))))...........(((....)))..)))).. ( -20.82) >DroYak_CAF1 86787 119 - 1 AGUUGCGACGCGCAGCUGCAGCACAAA-AAAUACAACAGCAACAACUACAGCUACAAAAUUGCAUGCACGAAAAUGCAAAAAAUCAAUAGCCAAACGAAAUUAGCCAAUGCUUUUUGCAC (((((((...)))))))(((((.....-.........(((..........)))......((((((........))))))..........))...........(((....)))...))).. ( -19.90) >consensus AGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGCAACAACUACAGCUACAAAAUUGCAUGCACGAAAAUGCAAAAAAUCAAUAGCCAAACGAAAUUAGCCAAUGCUUUCUGCAC (((((((...)))))))((((.............................((((.....((((((........))))))........))))...........(((....)))..)))).. (-20.56 = -20.37 + -0.19)

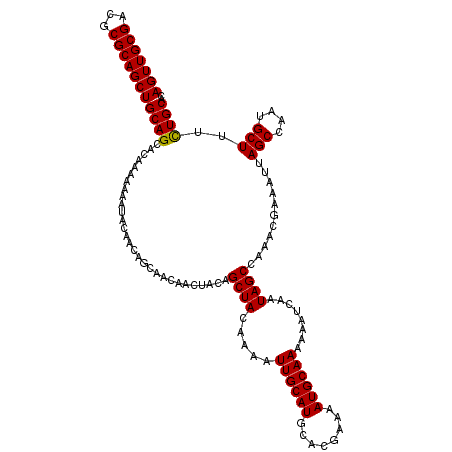
| Location | 5,942,032 – 5,942,147 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5942032 115 - 20766785 ACUCUCUCAAUCACUCGUGCACUCU-CUUUCGUUAACUGACCCAAAAUACAA----ACAACAACAGCAACAGCAGUUUACAGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGC ................((((.....-..........................----.........((....))......((((((((...))))))))..))))................ ( -18.60) >DroSec_CAF1 84114 119 - 1 ACGCUCUCACUCACUCGUGCACUCU-CUUUCGUUAACUGACCCAAAAUACAAAACAACAACAACAGCAACAGCAGUUUACAGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGC ................((((.....-.......................................((....))......((((((((...))))))))..))))................ ( -18.60) >DroSim_CAF1 81961 120 - 1 ACUCUCUCACUCACUCGUGCACUCUUCUUUCGUUAACUGACCCAAAAUACAAAACAACAACAACAGCAACAGCAGUUUACAGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGC ................((((.............................................((....))......((((((((...))))))))..))))................ ( -18.60) >DroYak_CAF1 86867 114 - 1 ACUCUCUCACUCACUCGUGCACUCU-CUUUUGUUAACUGACCCCAAAUAUAA-ACAA---CAACAGCAACAGCAGAUUACAGUUGCGACGCGCAGCUGCAGCACAAA-AAAUACAACAGC ................((((....(-((.(((((..(((.............-....---...))).))))).)))...((((((((...))))))))..))))...-............ ( -21.15) >consensus ACUCUCUCACUCACUCGUGCACUCU_CUUUCGUUAACUGACCCAAAAUACAA_ACAACAACAACAGCAACAGCAGUUUACAGUUGCGACGCGCAGCUGCAGCACAAAAAAAUACAACAGC ................((((.............................................((....))......((((((((...))))))))..))))................ (-18.60 = -18.60 + -0.00)

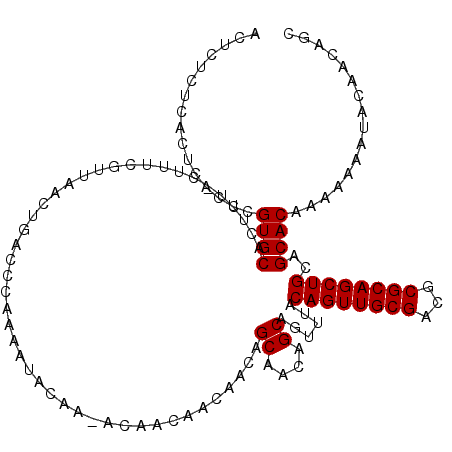
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:03 2006