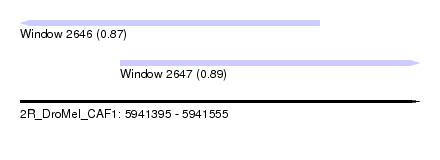
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,941,395 – 5,941,555 |
| Length | 160 |
| Max. P | 0.885783 |

| Location | 5,941,395 – 5,941,515 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5941395 120 - 20766785 UCGAUGAUAUCGAUAUCUAUUCCGACACCUGACAACAUCAGAGUGCGCUCGGCAGCUAUCGAUUAUUCUUAUCGAAUAAAUCUCUUUAGGCGCCUAAAUUCAAAAUUUGAUAAGCAGUAA ........(((((((.((...(((((((((((.....)))).)))...)))).)).)))))))((((((((((((((.......(((((....)))))......)))))))))).)))). ( -30.12) >DroSec_CAF1 83476 120 - 1 UCGAUGCUAUCGAUAUUUAUUCCGACACCUGACAACAUCCGAGUGCGCUCGCUAGCCAUCGAUUAUUCUUAUCGAAUAAAUCUCUUUAGGCGCCUAAGUGUGAAAUUUGAUAAGCAAUAA (((((((((.(((....(((((.((............)).)))))...))).))).))))))(((((((((((((((...((.((((((....))))).).)).)))))))))).))))) ( -28.10) >DroSim_CAF1 81324 120 - 1 UCGAUGCUAUCGAUAUUUAUUCCGACACCUGACAACAUCCGAGUGCGCUCCCUAGCUAUCGAUUAUUCUUAUCGAAUAAAUCUAUUUAGGCGCCUAAAUGUGAAAUUUGAUAAGCAAUAA ((((((((((((..........)))...............(((....)))..)))).)))))(((((((((((((((...(((((((((....))))))).)).)))))))))).))))) ( -27.70) >DroEre_CAF1 68622 106 - 1 UCGAUGCUAUCGAU--UCUAU------------CAGACACUAGUCUGCUCGGCAGCUAUCGAUUAUUCAUAUCGAUUAAAUUCCUCUUGGCGCCUAAAUUCAAAAUCUGAUAAGCAAUAG ....((((((((((--(....------------(((((....)))))...(((.(((((((((.......))))))............)))))).........)))).))).)))).... ( -23.20) >DroYak_CAF1 86245 106 - 1 UCGACGCUAUCGAU--UGAUU------------AACAUCAGAGUGUGCUUGGCAGCUAUCGAUAAUUCUUAUCGAUUAAGUCCCUCUUGGCGCUGAAAUGCAAAAUCUGAUAAGCAAUAC .....((((((...--.))).------------...((((((...(((....((((.(((((((.....)))))))...(((......)))))))....)))...)))))).)))..... ( -27.60) >consensus UCGAUGCUAUCGAUAUUUAUUCCGACACCUGACAACAUCCGAGUGCGCUCGGCAGCUAUCGAUUAUUCUUAUCGAAUAAAUCUCUUUAGGCGCCUAAAUGCAAAAUUUGAUAAGCAAUAA ........((((((.....................(((....))).(((....))).))))))((((((((((((((.......(((((....)))))......)))))))))).)))). (-13.18 = -13.78 + 0.60)

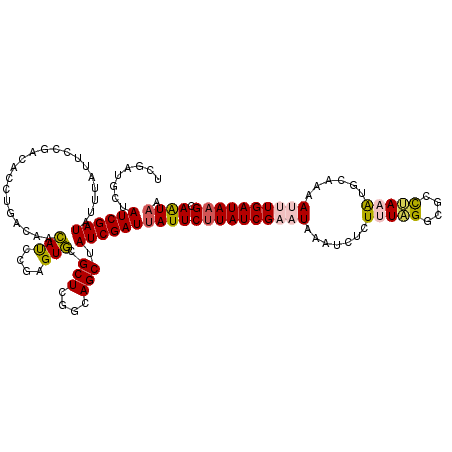
| Location | 5,941,435 – 5,941,555 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -19.42 |
| Energy contribution | -22.42 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5941435 120 + 20766785 UUUAUUCGAUAAGAAUAAUCGAUAGCUGCCGAGCGCACUCUGAUGUUGUCAGGUGUCGGAAUAGAUAUCGAUAUCAUCGAAUGAACAUCGGCUUUUGUUCCAGCUCUAUAGCAUAAAGUC .((((((.....))))))......((((..((((((((.(((((...))))))))).((((((((..(((((.((((...))))..)))))..)))))))).))))..))))........ ( -37.90) >DroSec_CAF1 83516 120 + 1 UUUAUUCGAUAAGAAUAAUCGAUGGCUAGCGAGCGCACUCGGAUGUUGUCAGGUGUCGGAAUAAAUAUCGAUAGCAUCGAAUGAACAUCGGUUUUUGUUCCAGCUCUAUAGCAUAAAGUC .((((((.....))))))......((((..((((((((..((......))..)))).((((((((.((((((..(((...)))...)))))).)))))))).))))..))))........ ( -34.80) >DroSim_CAF1 81364 120 + 1 UUUAUUCGAUAAGAAUAAUCGAUAGCUAGGGAGCGCACUCGGAUGUUGUCAGGUGUCGGAAUAAAUAUCGAUAGCAUCGAAUGAACAUCGGUUUUUGUUCCAGCUCUAUAGCAUAAAGUC .((((((.....))))))......((((.(((((((((..((......))..)))).((((((((.((((((..(((...)))...)))))).)))))))).))))).))))........ ( -37.20) >DroEre_CAF1 68662 106 + 1 UUUAAUCGAUAUGAAUAAUCGAUAGCUGCCGAGCAGACUAGUGUCUG------------AUAGA--AUCGAUAGCAUCGAAUAAACAUCGGUUUUUGUUCCACCUCUAUAGCAUAAAGUC ...(((((((.(...((.(((((.((((.(((.(((((....)))))------------.....--.))).))))))))).)).).)))))))........................... ( -23.20) >DroYak_CAF1 86285 106 + 1 CUUAAUCGAUAAGAAUUAUCGAUAGCUGCCAAGCACACUCUGAUGUU------------AAUCA--AUCGAUAGCGUCGAAUAAACAUCGCCUUUUGUUCCAGCUCUAUAGCAUAAAGUC ....(((((((.....))))))).((((...(((.......((((((------------(.((.--...)))))))))(((((((........)))))))..)))...))))........ ( -23.80) >consensus UUUAUUCGAUAAGAAUAAUCGAUAGCUGCCGAGCGCACUCGGAUGUUGUCAGGUGUCGGAAUAAAUAUCGAUAGCAUCGAAUGAACAUCGGUUUUUGUUCCAGCUCUAUAGCAUAAAGUC .....(((((.......)))))..((((..((((.......(((((((((.(((((........))))))))))))))(((((((........)))))))..))))..))))........ (-19.42 = -22.42 + 3.00)

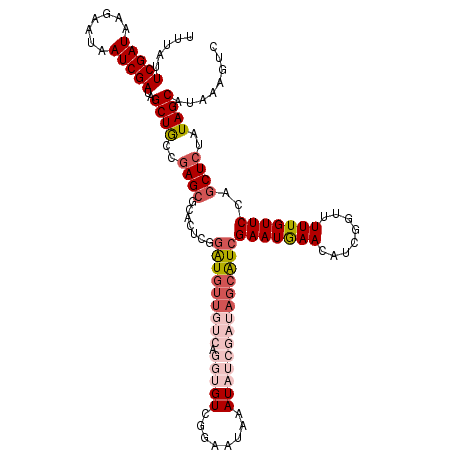
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:00 2006