
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
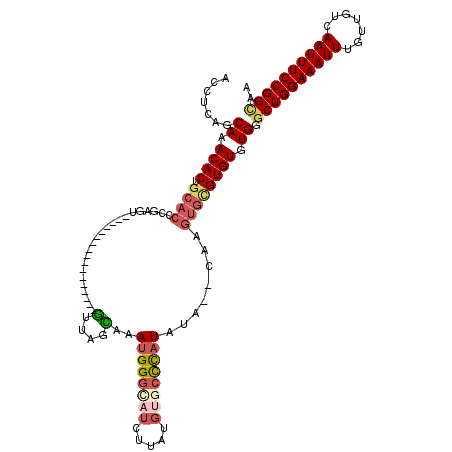
| Location | 5,908,856 – 5,908,957 |
| Length | 101 |
| Max. P | 0.661350 |

| Location | 5,908,856 – 5,908,957 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -22.25 |
| Energy contribution | -23.28 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
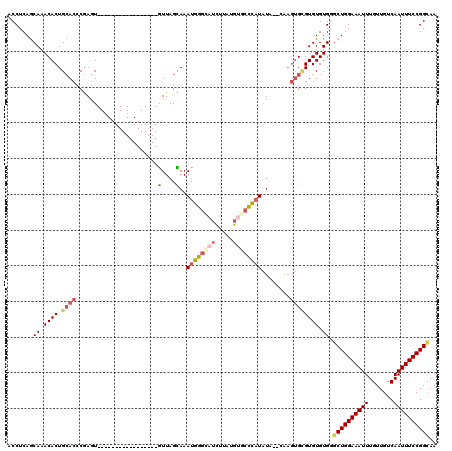
>2R_DroMel_CAF1 5908856 101 + 20766785 ACCUCAGCAAACACUGCACCCGAGU-----------------GUUAGCAAAUGGAGAUCUUAUGUGCCCAUAUA--CAAGUGCGUGUGUGGGCUGGAAAUUUGUUGUCAAUUUCCGGCAA ..(((.((.((((((.......)))-----------------))).)).....)))......((((((((((((--(......))))))))))((((((((.......))))))))))). ( -32.70) >DroSec_CAF1 50897 103 + 1 ACCUCAGCAAACACUGCACCCGAGU-----------------GUUAGCAAAUGGGCAUCUUAUGUGCCCAUAUACACAAGUGCGUGUGUGGGCUGGAAAUUUGUUGUCAAUUUCCGGUAA (((..(((..(((((((((....((-----------------((......((((((((.....))))))))..))))..))))).))))..)))(((((((.......)))))))))).. ( -37.50) >DroSim_CAF1 44091 103 + 1 ACCUCAGCAAACACUGCACCCGAGU-----------------GUUAGCAAAUGGGCAUCUUAUGUGCCCAUAUACGCAAGUGCGUGUGUGGGCUGGAAAUUUGUUGUCAAUUUCCGGCAA .((.(.((.((((((.......)))-----------------))).))..((((((((.....))))))))((((((....))))))).))((((((((((.......)))))))))).. ( -41.50) >DroEre_CAF1 43116 99 + 1 ACCUCAGCAAACACUGCACCCGAGU-----------------GUUAGUAAAUGGGCAUCUUAUGUGCCCUUAU----AAGUGCGUGUGUGGGCUGGAAAUUUGUUGUCAAUUUCCGGCAA .((...(((..((((((((....))-----------------))........((((((.....))))))....----.))))..)))..))((((((((((.......)))))))))).. ( -33.60) >DroYak_CAF1 43395 97 + 1 ACCUCAGCAAACACUGCACCCGAGU-----------------GUUAGUAAAUGGGUCUCUUAUGUGCCCAUAU------GUGUGUGUGUGUGCUGGAAAUUUGUUGUCAAUUUCCGGCAA ......(((.((((.((((....))-----------------))......(((((((......).))))))..------)))).)))...(((((((((((.......))))))))))). ( -28.80) >DroAna_CAF1 50358 120 + 1 ACCUCAACAAACACUGGACCCAAUUUUCCGUAUGUGUAGUUGAUUAGUAAAUGGGCUCUUUAUAUCCUUAUGUAAGCGACAGAGUGUGUGUCCUGGAAAUUUGUUGUCAAUUUCCGGCAA ...(((((..((((((((........))))...)))).))))).......(((.(((((((((((....)))))).....))))).)))...(((((((((.......)))))))))... ( -23.20) >consensus ACCUCAGCAAACACUGCACCCGAGU_________________GUUAGCAAAUGGGCAUCUUAUGUGCCCAUAUA__CAAGUGCGUGUGUGGGCUGGAAAUUUGUUGUCAAUUUCCGGCAA .......((.((((.((((.......................(....)..((((((((.....))))))))........)))))))).)).((((((((((.......)))))))))).. (-22.25 = -23.28 + 1.03)

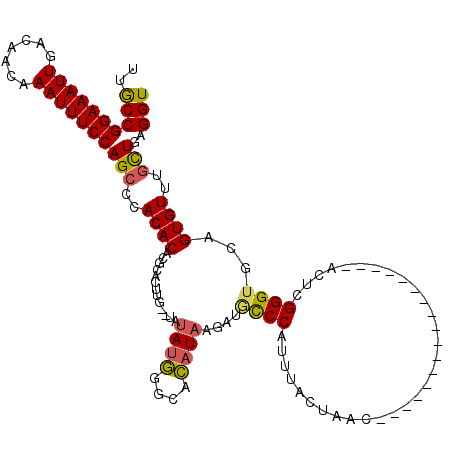
| Location | 5,908,856 – 5,908,957 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
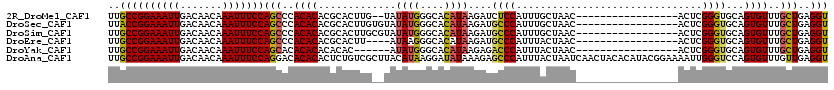
>2R_DroMel_CAF1 5908856 101 - 20766785 UUGCCGGAAAUUGACAACAAAUUUCCAGCCCACACACGCACUUG--UAUAUGGGCACAUAAGAUCUCCAUUUGCUAAC-----------------ACUCGGGUGCAGUGUUUGCUGAGGU ..((((((((((.......)))))))(((..((((..(((((((--......((((.((.........)).))))...-----------------...))))))).))))..)))..))) ( -29.00) >DroSec_CAF1 50897 103 - 1 UUACCGGAAAUUGACAACAAAUUUCCAGCCCACACACGCACUUGUGUAUAUGGGCACAUAAGAUGCCCAUUUGCUAAC-----------------ACUCGGGUGCAGUGUUUGCUGAGGU ..((((((((((.......)))))))(((..((((..(((((((.(((.((((((((....).))))))).)))....-----------------...))))))).))))..)))..))) ( -37.10) >DroSim_CAF1 44091 103 - 1 UUGCCGGAAAUUGACAACAAAUUUCCAGCCCACACACGCACUUGCGUAUAUGGGCACAUAAGAUGCCCAUUUGCUAAC-----------------ACUCGGGUGCAGUGUUUGCUGAGGU ..((((((((((.......)))))))(((..((((..(((((((.(((.((((((((....).))))))).)))....-----------------...))))))).))))..)))..))) ( -36.00) >DroEre_CAF1 43116 99 - 1 UUGCCGGAAAUUGACAACAAAUUUCCAGCCCACACACGCACUU----AUAAGGGCACAUAAGAUGCCCAUUUACUAAC-----------------ACUCGGGUGCAGUGUUUGCUGAGGU ..((((((((((.......)))))))(((..((((..(((((.----....((((((....).))))).........(-----------------....)))))).))))..)))..))) ( -29.20) >DroYak_CAF1 43395 97 - 1 UUGCCGGAAAUUGACAACAAAUUUCCAGCACACACACACAC------AUAUGGGCACAUAAGAGACCCAUUUACUAAC-----------------ACUCGGGUGCAGUGUUUGCUGAGGU ..((((((((((.......)))))))((((.((((......------..((((((........).))))).......(-----------------((....)))..)))).))))..))) ( -26.50) >DroAna_CAF1 50358 120 - 1 UUGCCGGAAAUUGACAACAAAUUUCCAGGACACACACUCUGUCGCUUACAUAAGGAUAUAAAGAGCCCAUUUACUAAUCAACUACACAUACGGAAAAUUGGGUCCAGUGUUUGUUGAGGU ..((((((((((.......)))))))..((((.(((((.((((.(((....)))))))....(..((((.............................))))..)))))).))))..))) ( -26.65) >consensus UUGCCGGAAAUUGACAACAAAUUUCCAGCCCACACACGCACUUG__UAUAUGGGCACAUAAGAUGCCCAUUUACUAAC_________________ACUCGGGUGCAGUGUUUGCUGAGGU ..((((((((((.......)))))))(((..((((.............((((....))))....((((...............................))))...))))..)))..))) (-19.42 = -19.28 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:39 2006