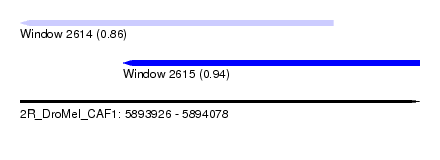
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,893,926 – 5,894,078 |
| Length | 152 |
| Max. P | 0.940245 |

| Location | 5,893,926 – 5,894,045 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -34.58 |
| Energy contribution | -34.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5893926 119 - 20766785 GUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGGCCGCCAUAAACAAGCGUUAAUUGCACUUUGAGGUUGCAC ((..(((((((((.(((....(((.....(((.(((((((.((.((.((.(....).)))))).))))))))))))).))))))))))))...))......((((((....)).)))). ( -39.80) >DroSec_CAF1 30180 119 - 1 GUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCCCAGUGAAUUGAGGCUGGGCCGCCAUAAACAAGCGUUAAUUGCACUUUGAGGUUGCAC ((..(((((((((.(((....(((.....(((.(((((((........((((........))))))))))))))))).))))))))))))...))......((((((....)).)))). ( -36.90) >DroSim_CAF1 28940 119 - 1 GUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGGCCGCCAUAAACAAGCGUUAAUUGCACUUUGAGGUUGCAC ((..(((((((((.(((....(((.....(((.(((((((.((.((.((.(....).)))))).))))))))))))).))))))))))))...))......((((((....)).)))). ( -39.80) >DroEre_CAF1 28237 119 - 1 GUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUUACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGGCCGCCAUAAACAAGCGUUAAUUGCACUUUGAGGUUGCAC ((..(((((((((.(((....(((.....(((.(((((((.((.((.((.(....).)))))).))))))))))))).))))))))))))...))......((((((....)).)))). ( -37.60) >DroYak_CAF1 28271 119 - 1 GUGAUUUGCGGCGAGCCGCAAAUCUGAAGCUCGAAUUUACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGGCAGCCAUAAACAAGCGUUAAUUGCACUGUGAAGUUGCAC ..(((((((((....)))))))))...((((((..........))))))..(..((...((((((((((((((.(((.(...........).))).)))))).)))))))).))..).. ( -43.30) >consensus GUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGGCCGCCAUAAACAAGCGUUAAUUGCACUUUGAGGUUGCAC ((..(((((((((.(((....(((.....(((.(((((((.((.((.((.(....).)))))).))))))))))))).))))))))))))...))......((((((....)).)))). (-34.58 = -34.98 + 0.40)

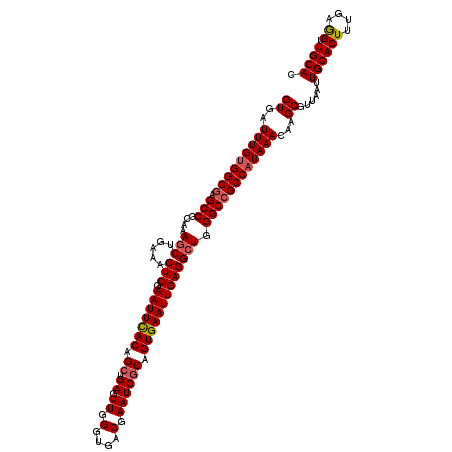
| Location | 5,893,965 – 5,894,078 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.93 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5893965 113 - 20766785 CUGCUCGGCGUGAAUUUAAUUACAGCCCCAUAAGUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGG ...((((((.............((((..((....)).(((((((....)))))))))))...(((.(((((((.((.((.((.(....).)))))).)))))))))))))))) ( -36.90) >DroSec_CAF1 30219 113 - 1 CUGCUCGGCGUGAAUUUAAUUACAGCCCCAUAAGUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCCCAGUGAAUUGAGGCUGGG (.((((((((((((((..(((.((((..((....)).(((((((....))))))))))).)).)..))))))).))))))).)(....)....((((((........)))))) ( -40.50) >DroSim_CAF1 28979 113 - 1 CUGCUCGGCGUGAAUUUAAUUACAGCCCCAUAAGUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGG ...((((((.............((((..((....)).(((((((....)))))))))))...(((.(((((((.((.((.((.(....).)))))).)))))))))))))))) ( -36.90) >DroEre_CAF1 28276 113 - 1 CUGCUCGGUAUGAAUUUAAUUGCAGCUCCAUAAGUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUUACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGG ...((((((.(.((((((.(((((((((..........((((((....))))))((((((((......))).)))))))))).(....).....)))))))))).).)))))) ( -33.40) >DroYak_CAF1 28310 113 - 1 CUGCUUGGCAUGAAUUUAAUUACAGCUCCAUAAGUGAUUUGCGGCGAGCCGCAAAUCUGAAGCUCGAAUUUACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGG ...((..((.(.((((((....((((((.......(((((((((....)))))))))...((((.(......))))))))))).((((....))))..)))))).).))..)) ( -36.40) >consensus CUGCUCGGCGUGAAUUUAAUUACAGCCCCAUAAGUGAUUUGUGGCGAGCCGCAAAGCUGAAACUCGAAUUCACAGCUGAGUUGGGUGACGAAUCGCAGUGAAUUGAGGCUGGG ...((((((.............((((..((....)).(((((((....)))))))))))...(((.(((((((.((.((.((.(....).)))))).)))))))))))))))) (-33.56 = -33.24 + -0.32)

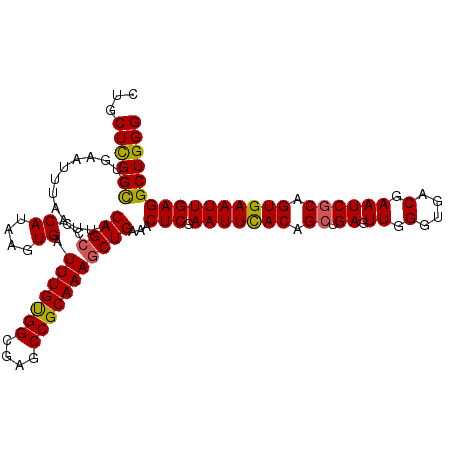
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:28 2006