
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,888,682 – 5,888,837 |
| Length | 155 |
| Max. P | 0.742244 |

| Location | 5,888,682 – 5,888,797 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -30.18 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
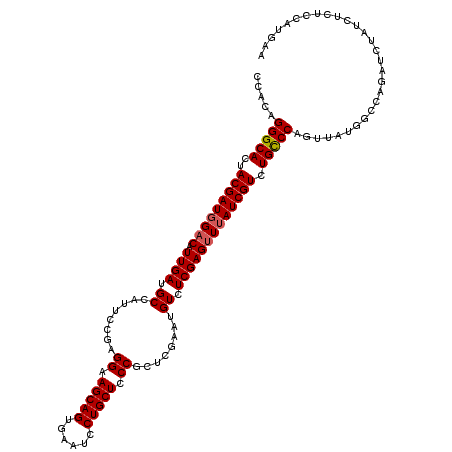
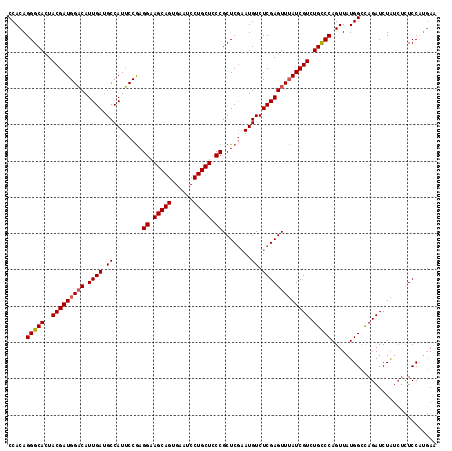
>2R_DroMel_CAF1 5888682 115 + 20766785 CCACAGGGCACUACGAUGGCCAUUGA-GCCAUGCCAAGGAAGCAGUGAAUCCUGCUACCGCUCAAAUGUCUCGAGUUGAUCGUCUGCCCAGUUAUGGGUAGAUCUGGG--UCCUUUAU ....((((((((.(((.(((..((((-((........((.(((((......))))).))))))))..))))))))).....(((((((((....)))))))))....)--)))).... ( -40.80) >DroSec_CAF1 24826 118 + 1 CCACAGGGCACUACGAUGGACAUUGAUGCCAUUCCGAGGAAGCAGUGAAUCCUGCUCCCGCUCGAAUGUCUCGAGUUUAUCGUCUGCCCAGUUAUGGCCAGAUCUAUCUCUCCCCGAA .....(((((..(((((((((.((((.(.(((((((.(((.((((......)))))))))...))))).)))))))))))))).))))).....(((..(((......)))..))).. ( -39.90) >DroSim_CAF1 23471 118 + 1 CCACAGGGCACUACGAUGGACAUUGAUGCCAUUCCGAGGAAGCAGUGAAUCCUGCUCCCGCUCGCAUGUCUCGAGUUUAUCGUCUGUCCAGUUAUGGCCAGAUCUAUCUCUCCAUGAA .....(((((..(((((((((.((((.(.(((..(((((.(((((......)))))..).)))).))).)))))))))))))).)))))..((((((..(((....)))..)))))). ( -38.30) >consensus CCACAGGGCACUACGAUGGACAUUGAUGCCAUUCCGAGGAAGCAGUGAAUCCUGCUCCCGCUCGAAUGUCUCGAGUUUAUCGUCUGCCCAGUUAUGGCCAGAUCUAUCUCUCCAUGAA .....(((((..(((((((((.((((.((........((.(((((......))))).))........)).))))))))))))).)))))............................. (-30.18 = -30.62 + 0.45)

| Location | 5,888,721 – 5,888,837 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.53 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
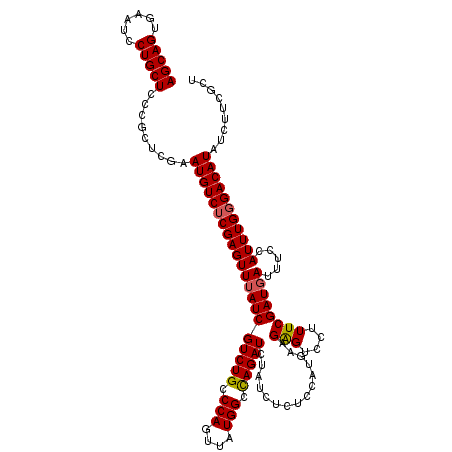
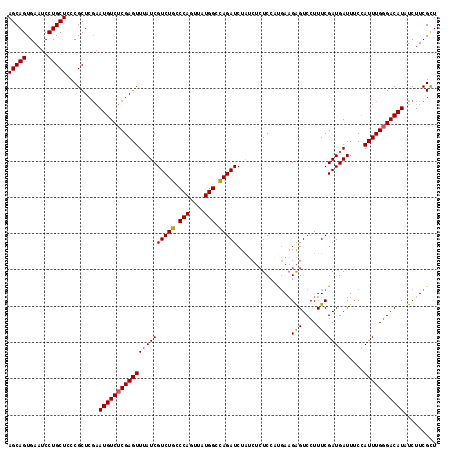
>2R_DroMel_CAF1 5888721 116 + 20766785 AGCAGUGAAUCCUGCUACCGCUCAAAUGUCUCGAGUUGAUCGUCUGCCCAGUUAUGGGUAGAUCUGGG--UCCUUUAUGGGUCCUUUCGAUGAUUACCAUUUGCGACAUAUCUUCGUU (((((......))))).........(((((.(((((((((((((((((((....))))))((...((.--.((.....))..))..))))))))))..))))).)))))......... ( -40.50) >DroSec_CAF1 24866 118 + 1 AGCAGUGAAUCCUGCUCCCGCUCGAAUGUCUCGAGUUUAUCGUCUGCCCAGUUAUGGCCAGAUCUAUCUCUCCCCGAAGAGUCCUUUCGAUGAUUUCCAUUUGGGACAUUUCUUCGCU (((((......)))))...((((((.....)))))).....(((((.(((....))).)))))...........(((((((((((...((((.....)))).)))))...)))))).. ( -34.10) >DroSim_CAF1 23511 118 + 1 AGCAGUGAAUCCUGCUCCCGCUCGCAUGUCUCGAGUUUAUCGUCUGUCCAGUUAUGGCCAGAUCUAUCUCUCCAUGAAGAGUCCUUUCGAUGAUUUCUAUUUGGGACAUAUCUUCGCU ...((((((....((........))(((((((((((..((((((.......((((((..(((....)))..)))))).(((....)))))))))....)))))))))))...)))))) ( -31.80) >consensus AGCAGUGAAUCCUGCUCCCGCUCGAAUGUCUCGAGUUUAUCGUCUGCCCAGUUAUGGCCAGAUCUAUCUCUCCAUGAAGAGUCCUUUCGAUGAUUUCCAUUUGGGACAUAUCUUCGCU (((((......))))).........(((((((((((((((((((((.(((....))).)))))...............(((....)))))))).....)))))))))))......... (-25.31 = -25.53 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:24 2006