| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,879,408 – 5,879,507 |
| Length | 99 |
| Max. P | 0.929271 |

| Location | 5,879,408 – 5,879,507 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.17 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
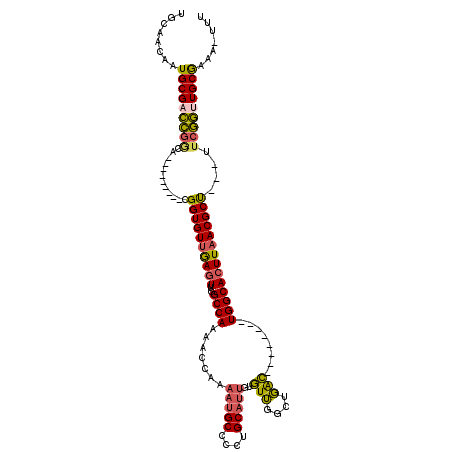
>2R_DroMel_CAF1 5879408 99 + 20766785 UGCAACAAUGCGACCGGCA---------UGGUGUUGAGUUGGCCAAAACCAAAAUGCCCCUGCAUUGUGUUGGCUGAC--------UGGCACUUAACGCU---UUCGGUUGCGAAA-UUU ........((((((((((.---------.((((((.(((..(((((......(((((....)))))...)))))..))--------)))))))....)).---..))))))))...-... ( -36.90) >DroSec_CAF1 15475 99 + 1 UGCAACAAUGCGACCGUCA---------CGGUGUUGAGUUGGCCAAAACCAAAAUGCCCCUGCAUUGUGUUGCCUGAC--------UGGCACUUAACGCU---UUCGGUUGCGAAA-UUU ........((((((((...---------.(((((((((((((......))))(((((....)))))....((((....--------.)))))))))))))---..))))))))...-... ( -33.70) >DroSim_CAF1 14090 99 + 1 UGCAACAAUGCGACCGGCA---------CGGUGUUGAGUUGGCCAAAACCAAAAUGCCCCUGCAUUGUGUUGCCUGAC--------UGGCACUUAACGCU---UUCGGUUGCGAAA-UUU ........(((((((((..---------.(((((((((((((......))))(((((....)))))....((((....--------.)))))))))))))---.)))))))))...-... ( -34.10) >DroEre_CAF1 15330 99 + 1 UGCAACAAUGCGACCGGCG---------CGGUGUUGAGUUGGCCAAAACCAAAAUGCCCCUGCAUUGUGUUCGCUGAC--------UGGCACUUAACGCU---UUCGGUUGCAAAA-UUU ........(((((((((((---------.((((((.(((..((...(((...(((((....)))))..))).))..))--------)))))))...))).---..))))))))...-... ( -34.20) >DroYak_CAF1 14829 107 + 1 UGCAACAAUGCGACCGACA---------CGGUGUUGAGUUGGCCAAAACCAAAAUGCCCCUGCAUUGUGUUGGCUGACUUUUGUGUUGGCACUUAACGCU---UUCGGCUGCGAAA-UUU .(((....)))(.((((((---------(((....((((..(((((......(((((....)))))...)))))..))))))))))))))......(((.---.......)))...-... ( -35.50) >DroPer_CAF1 17364 108 + 1 UGCAACAAUGCGUAGUGCGGGGGGGCGGGGGUGUUAACUUGGCCAACUGC----UGCUUCUGCCACUUCUGUGCUCGG--------UGGCAUUUCACGCCGCACUCCUUUGCGGGGUUUU .(((....)))(.((..(((((((((((((((........(((.....))----)))))))))).))))))..))).(--------((((.......)))))(((((.....)))))... ( -41.10) >consensus UGCAACAAUGCGACCGGCA_________CGGUGUUGAGUUGGCCAAAACCAAAAUGCCCCUGCAUUGUGUUGGCUGAC________UGGCACUUAACGCU___UUCGGUUGCGAAA_UUU ........(((((((((............((((((((((..((((.......(((((....)))))..(((....)))........))))))))))))))....)))))))))....... (-26.44 = -26.17 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:20 2006