| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,820,012 – 5,820,120 |
| Length | 108 |
| Max. P | 0.829198 |

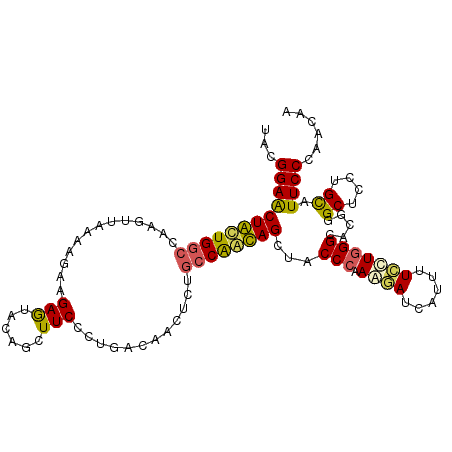
| Location | 5,820,012 – 5,820,120 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -22.81 |
| Energy contribution | -20.62 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
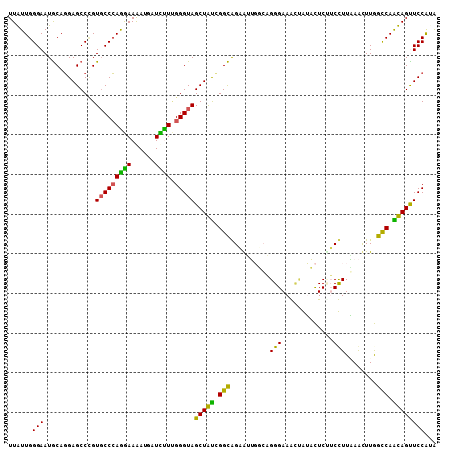
>2R_DroMel_CAF1 5820012 108 + 20766785 UUAUUGGGAAUGCAGGAGCCUGUGCCCAAGAAAAUUAUCUUCGGGUAGCUAUCGGCAGAAUUGUCAGGGAAACUAUACUCUUCUUUGAAUUUGGCCAAUAGUUCCAGA ...(((((...((((....)))).)))))..............((.((((((.(((.((((..((((((((.........)))))))))))).))).))))))))... ( -32.20) >DroPse_CAF1 18342 108 + 1 UUGUUGGGAAUGCAGGAGCCCGUGCCCAGGAAGAUGAUCCUUGGGUAGCUGCUGGCGGCGGACGAUGGGAAGUUGGUUUCGGCCUUUAGCUUAGCCAGCAGCUCCGUA ..............(((((((((((((((((......))).))))).((((....)))).....))))...(((((((..(((.....))).))))))).)))))... ( -40.50) >DroSec_CAF1 17512 108 + 1 UUAUUGGGAAUGCAGGAGCCAGUGCCCAAGAAAAUGAUCUUUGGGUAGCUAUCGGCAGAAUUGUCAGGGAAACUAUACUCUUCUUUGAACUUGGCCAAUAGUUCCAGA ......(((((....(.((((((((((((((......)).)))))))....((((.((((..((..((....))..))..))))))))..))))))....)))))... ( -30.50) >DroYak_CAF1 17710 108 + 1 UUAUUGGGAAUGCAGGAGCCUGUGCCCAGGAAAAUGAUUUUCGGGUAGCUAUCGGCAGUAUUGUCAGGGAAACUAUACUCAUCUUUAAAUUUGGUCAAUAGUUCCAUA ...(((((...((((....)))).)))))((((.....)))).((.((((((.((((((((.....((....)))))))((..........))))).))))))))... ( -25.00) >DroAna_CAF1 15078 108 + 1 UUGUUGGGAAUACACGAGCCCGUUCCGAGAAAAAUAAUUUUGGGGUAGCUGUUGGUAGAGCUGGCCGGGAAGGAGUACUCUUCCUUAAACUUGGCCAGCAGUUCCGUG ......(((((.((..(((..(..((((((.......))))))..).)))..)).....((((((((((((((((.....))))))...)))))))))).)))))... ( -39.30) >DroPer_CAF1 18855 108 + 1 UUGUUGGGAAUGCAGGAGCCCGUGCCCAGGAAGAUGAUCCUUGGGUAGCUGCUGGCGGCGGACGAUGGGAAGUCGGUUUCCGCCUUUAGCUUAGCCAGCAGCUCCGUA ..............(((.....(((((((((......))).))))))(((((((((((((((((((.....))))...)))))).........))))))))))))... ( -46.00) >consensus UUAUUGGGAAUGCAGGAGCCCGUGCCCAGGAAAAUGAUCUUUGGGUAGCUAUCGGCAGAAUUGGCAGGGAAACUAUACUCUUCCUUAAACUUGGCCAACAGUUCCAUA ......(((.............(((((((((......)))).)))))(((((.(((..........(((.............)))........))).))))))))... (-22.81 = -20.62 + -2.19)

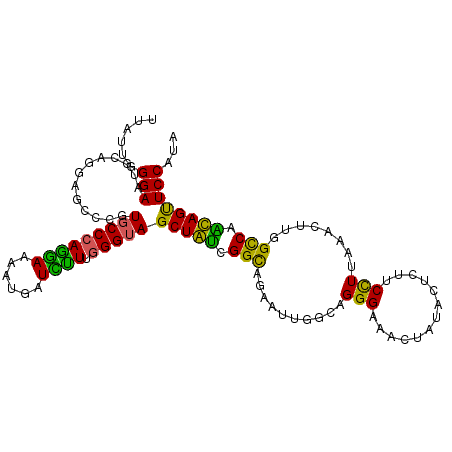
| Location | 5,820,012 – 5,820,120 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -18.85 |
| Energy contribution | -17.91 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5820012 108 - 20766785 UCUGGAACUAUUGGCCAAAUUCAAAGAAGAGUAUAGUUUCCCUGACAAUUCUGCCGAUAGCUACCCGAAGAUAAUUUUCUUGGGCACAGGCUCCUGCAUUCCCAAUAA ...((((((((((((...((((......))))...(((.....)))......))))))))...(((.((((......)))))))..(((....)))..))))...... ( -24.60) >DroPse_CAF1 18342 108 - 1 UACGGAGCUGCUGGCUAAGCUAAAGGCCGAAACCAACUUCCCAUCGUCCGCCGCCAGCAGCUACCCAAGGAUCAUCUUCCUGGGCACGGGCUCCUGCAUUCCCAACAA ...((((((((((((...((....((.(((.............))).)))).)))))))))).((((.(((......)))))))..(((....))).....))..... ( -35.62) >DroSec_CAF1 17512 108 - 1 UCUGGAACUAUUGGCCAAGUUCAAAGAAGAGUAUAGUUUCCCUGACAAUUCUGCCGAUAGCUACCCAAAGAUCAUUUUCUUGGGCACUGGCUCCUGCAUUCCCAAUAA ...((((((((((((..((((((..((((.......))))..)))..)))..))))))))...(((((.((......))))))).....((....)).))))...... ( -23.50) >DroYak_CAF1 17710 108 - 1 UAUGGAACUAUUGACCAAAUUUAAAGAUGAGUAUAGUUUCCCUGACAAUACUGCCGAUAGCUACCCGAAAAUCAUUUUCCUGGGCACAGGCUCCUGCAUUCCCAAUAA ...((((((((((.....(((....))).(((((.(((.....))).)))))..))))))......(.....)...))))((((..(((....)))....)))).... ( -17.20) >DroAna_CAF1 15078 108 - 1 CACGGAACUGCUGGCCAAGUUUAAGGAAGAGUACUCCUUCCCGGCCAGCUCUACCAACAGCUACCCCAAAAUUAUUUUUCUCGGAACGGGCUCGUGUAUUCCCAACAA (((((....(((((((........(((((.......))))).)))))))....))...((((.((..((((....))))...))....)))).)))............ ( -25.60) >DroPer_CAF1 18855 108 - 1 UACGGAGCUGCUGGCUAAGCUAAAGGCGGAAACCGACUUCCCAUCGUCCGCCGCCAGCAGCUACCCAAGGAUCAUCUUCCUGGGCACGGGCUCCUGCAUUCCCAACAA ...((((((((((((.........((((((...(((.......))))))))))))))))))).((((.(((......)))))))..(((....))).....))..... ( -42.60) >consensus UACGGAACUACUGGCCAAGUUAAAAGAAGAGUACAGCUUCCCUGACAACUCUGCCAACAGCUACCCAAAGAUCAUUUUCCUGGGCACGGGCUCCUGCAUUCCCAACAA ...((((((((((((.............(((......)))............))))))))...(((.((((......))))))).....((....)).))))...... (-18.85 = -17.91 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:08 2006