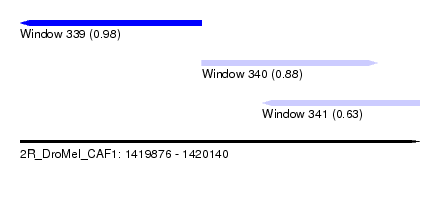
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,419,876 – 1,420,140 |
| Length | 264 |
| Max. P | 0.981384 |

| Location | 1,419,876 – 1,419,996 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1419876 120 - 20766785 GCCGGCGGCCGAACUGCAAUUUCAUUUCGCAUUCAUAUGCGAAAGUUUGGCGGAUGCUUUCAGCUGGCUUUCAAACAAAGGCGGUUUAAAUCGCCAUUAAAACUCGCUCACAUCCAGGAG ((((((.((((((((..........(((((((....)))))))))))))))(((....))).))))))...........((((((....)))))).......(((.((.......))))) ( -39.70) >DroSec_CAF1 38321 120 - 1 GCCGGCGGCCGAACUGCAAUUUCAUUUCGCAUUCAUAUGCGAAAGUUUGGCGCAUGCUUUCAGCUGGCUUUCAAACAAAGGCGGUUUAAAACGCCAUUAAAACUCGCUCACAUCCAGGAG ((((((.((((((((..........(((((((....)))))))))))))))(....).....))))))...........((((........)))).......(((.((.......))))) ( -36.90) >DroSim_CAF1 43376 120 - 1 GCCGGCGGCCGAACUGCAAUUUCAUUUCGCAUUCAUAUGCGAAAGUUUGGCGCAUGCUUUCAGCUGGCUUUCAAACAAAGGCGGUUUAAAUCGCCAUUAAAACUCGCUCACAUCCAGGAG ((((((.((((((((..........(((((((....)))))))))))))))(....).....))))))...........((((((....)))))).......(((.((.......))))) ( -37.70) >DroEre_CAF1 36798 120 - 1 GCCGGCGGCCGAACUGCAAUUUCAUUUCGCAUUCAUAUGCGAAAGUUUGGCGCAUGCUUUCAGCUCGCUUUCAAACAAAGGCGGUUUAAAGCGCCAUUAAAACUCGCUCACAUCCAUGAG ...(((.((((((((..........(((((((....)))))))))))))))....(((((.(((.((((((......))))))))).))))))))...........((((......)))) ( -39.60) >DroYak_CAF1 37637 120 - 1 GCCGACGGCAGAACUGCAAUAUAAUUACGCAUUCAUAUGCGAAAGUUUGGCGCAUGCUUUCAGCUCGCUUUCAAACAAAGGCAGUUUAAAUCGCCAUUAAAACUCGCUCACAUCCAUGAG ......(((.(((((((..........(((((....)))))...((((((.((..((.....))..))..))))))....))))))).....)))...........((((......)))) ( -29.00) >consensus GCCGGCGGCCGAACUGCAAUUUCAUUUCGCAUUCAUAUGCGAAAGUUUGGCGCAUGCUUUCAGCUGGCUUUCAAACAAAGGCGGUUUAAAUCGCCAUUAAAACUCGCUCACAUCCAGGAG ...((((...(((((((.......((((((((....))))))))((((((.((..((.....))..))..))))))....)))))))....))))...........(((........))) (-30.88 = -31.32 + 0.44)

| Location | 1,419,996 – 1,420,112 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -28.73 |
| Energy contribution | -30.42 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1419996 116 + 20766785 GCGUAUAUCUCGUGCGAAAGUUUGUCCAUUCCGACAUAAAUUGGGAUGGACCCUUAAAGCUGUGCACUAACAUUGGGGAGUAAGUAUCUAUCAGAUACCUUCCCCUU----CCUGUGGAG ........((((((((..((((((((((((((((......))))))))))).....))))).)))))...(((.((((((...(((((.....))))).))))))..----...)))))) ( -39.90) >DroSec_CAF1 38441 120 + 1 GCGUAUAUCUCGUGCGAAAGUUUGUCCAUCCCGACAUAAAUUGGGAUGGACCCUUAAAGCUGUGCACUAGCAUUGGGGAGCAAGUAUCUAUCAGAUACCUUCCCCGUCUCCCCUGUGGAG ........((((((((..((((((((((((((((......))))))))))).....))))).)))))...(((.((((((((((((((.....)))).)))....).)))))).)))))) ( -44.30) >DroSim_CAF1 43496 120 + 1 GCGUAUAUCUCGUGCGAAAGUUUGUCCAUCCCGACAUAAAUUGGGAUGGACCCUUAAAGCUGUGCACUAGCAUUGGGGAGCAAGUAUCUAUCAGAUACCUUCCCCGUCUCCCCUGUGGAG ........((((((((..((((((((((((((((......))))))))))).....))))).)))))...(((.((((((((((((((.....)))).)))....).)))))).)))))) ( -44.30) >DroEre_CAF1 36918 108 + 1 GCGUAUAUCUCCGGCGAAAGUUUGUCUAU-CCGACAUAAAUUGGCGUGGCCCCUUGAAGCUGUGCACUAGCAUUGG-GAACAAGUAUCUUUCGGAUACCUUC----------CUGUGGAG ........(((((((...(((.((((...-..)))).......(((..((........))..)))))).))...((-(((...(((((.....))))).)))----------)).))))) ( -27.70) >DroYak_CAF1 37757 110 + 1 GCGUAUAACUCCGGCGAAAGCUUGUCUAUUCCGCCAUAAAUUGGGAUGGACCCUUAAAGCUCUGCACUAACAUUGGGGAACAAGUAUCUAUCAGAUACCUUG----------CUGUGGAG ........(((((((....))).((((((((((........))))))))))......((((((.((.......)).)))....(((((.....)))))...)----------))..)))) ( -30.70) >consensus GCGUAUAUCUCGUGCGAAAGUUUGUCCAUCCCGACAUAAAUUGGGAUGGACCCUUAAAGCUGUGCACUAGCAUUGGGGAGCAAGUAUCUAUCAGAUACCUUCCCC_U____CCUGUGGAG ........((((((((..((((((((((((((((......))))))))))).....))))).))))).......((((((...(((((.....))))).))))))............))) (-28.73 = -30.42 + 1.69)

| Location | 1,420,036 – 1,420,140 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -18.49 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
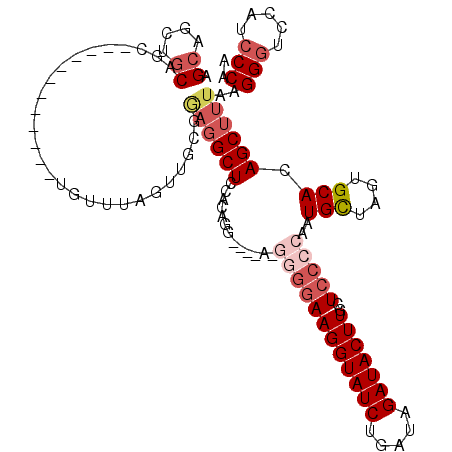
>2R_DroMel_CAF1 1420036 104 - 20766785 AGCAGCUGCAGC------------UGUUUAGUUGCGAAGGCUCCACAGG----AAGGGGAAGGUAUCUGAUAGAUACUUACUCCCCAAUGUUAGUGCACAGCUUUAAGGGUCCAUCCCAA .(((((((....------------....))))))).((((((.(((..(----..((((((((((((.....)))))))..)))))....)..)))...))))))..(((.....))).. ( -34.70) >DroSec_CAF1 38481 108 - 1 AGCAGCUGCAGC------------UGUUUAGUUGCGGAGGCUCCACAGGGGAGACGGGGAAGGUAUCUGAUAGAUACUUGCUCCCCAAUGCUAGUGCACAGCUUUAAGGGUCCAUCCCAA (((..(((((((------------(....))))))))..)))......(((((((((((((((((((.....)))))))..)))))..(((....)))...........)))..)))).. ( -42.20) >DroSim_CAF1 43536 108 - 1 AGCUGCUGCAGC------------UGUUUAGUUGCGGAGGCUCCACAGGGGAGACGGGGAAGGUAUCUGAUAGAUACUUGCUCCCCAAUGCUAGUGCACAGCUUUAAGGGUCCAUCCCAA ((((((((((((------------(....)))))))).(.((((.....)))).)((((((((((((.....)))))))..)))))..(((....))))))))....(((.....))).. ( -42.90) >DroEre_CAF1 36957 83 - 1 AGCAGCUGCAGCU--------------------------GCUCCACAG----------GAAGGUAUCCGAAAGAUACUUGUUC-CCAAUGCUAGUGCACAGCUUCAAGGGGCCACGCCAA ((((((....)))--------------------------)))((...(----------((((((((.(....))))))..)))-)...(((....))).........))(((...))).. ( -24.40) >DroYak_CAF1 37797 109 - 1 AGCGGCCACAGCUGGACCUGCAGCUGUUCAG-UGCGGAGGCUCCACAG----------CAAGGUAUCUGAUAGAUACUUGUUCCCCAAUGUUAGUGCAGAGCUUUAAGGGUCCAUCCCAA .(((...(((((((......)))))))....-))).((((((((((((----------(((((((((.....))))))).........)))).)))..)))))))..(((.....))).. ( -34.50) >consensus AGCAGCUGCAGC____________UGUUUAGUUGCGGAGGCUCCACAGG____A_GGGGAAGGUAUCUGAUAGAUACUUGCUCCCCAAUGCUAGUGCACAGCUUUAAGGGUCCAUCCCAA .((....))...........................((((((.............((((((((((((.....)))))))..)))))..(((....))).))))))..(((.....))).. (-18.49 = -20.38 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:16 2006