
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,807,768 – 5,807,881 |
| Length | 113 |
| Max. P | 0.547655 |

| Location | 5,807,768 – 5,807,881 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -41.22 |
| Consensus MFE | -26.33 |
| Energy contribution | -26.03 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5807768 113 - 20766785 UAACCAAAUUU-----UGGCACUUACCACGGGGUUUCUCGGGCGCUGGUUUACGGAAGCCGAGCAGAGGUGGCUCUCCCCCUGCCUGGAGACUCUUGUAAGCCUGCUUUUGUUCAACG ....((((...-----.(((((((((...((((((((.(((((((((((((....))))).)))..(((.((.....)))))))))))))))))).)))))..))))))))....... ( -38.60) >DroVir_CAF1 3729 109 - 1 ---C-AAAGUU-----GAGCUCUUACCUUUGGGUUUCUCGGGCGCGGGCUUACGGAAGCCCAGCAAAGGCGGUUCCCCUCCCGCCUGUAAGCUCUUGUAGGCCUGUCGCUGCUCCACG ---.-...((.-----((((....(((....)))......((((((((((((((..(((.......((((((........))))))....)))..)))))))))).)))))))).)). ( -45.70) >DroGri_CAF1 4368 109 - 1 ---U-GAGGUU-----UGAGGCUUACCUUUGGGUUUCUCGGGCGCUGGCUUACGGAAGCCGAGCAACGGCGGUUCACCUCCUGCCUGUAAGCUCUUGUAGGCCUGUCGCUGUUCCACG ---.-..(((.-----..((((((((....((((((..(((((((((((((....))))).)))...((.((....)).)).))))).))))))..))))))))...)))........ ( -42.10) >DroWil_CAF1 11777 107 - 1 -AUA-AAC---------GACUCUUACCUUUGGGUUUCUCGGGUGCUGGCUUACGGAAGCCAAGCAAGGGCGGUUCUCCACCGGCCUGUAGGCUUUUGUAGGCUUGCUUCUGCUCAACA -...-...---------(((((........)))))...((((.((.((((((((((((((..(((.((.((((.....)))).))))).)))))))))))))).)).))))....... ( -42.20) >DroAna_CAF1 3383 105 - 1 -AAG-ACA-----------GACUCACCACGGGGCUUCUCGGGCGCGGGCUUACGGAAGCCGAGCAGUGGAGGUUCUCCGCCUGCCUGGAGACUCUUGUAGGCCUGCCUUUGUUCCACG -..(-(((-----------((....((.((((....)))))).(((((((((((((..(((.(((((((((...))))).)))).)))....)).))))))))))).))))))..... ( -42.00) >DroPer_CAF1 3724 116 - 1 -AAG-AAAGACUCUCUCUUCACUUACCUCUGGGCCUCUCGGGCGCUGGCUUGCGGAAGCCGAAAAGGGGCGGUUCUCCGCCUGCCUGUAGACUCUUGUAGGCCUGUCGCUGCUCCACG -..(-((((.....)).)))..........(((((((((((((..((((((....)))))).....((((((....))))))))))).))((....)))))))).............. ( -36.70) >consensus _AAC_AAAGUU______GACACUUACCUCUGGGUUUCUCGGGCGCUGGCUUACGGAAGCCGAGCAAAGGCGGUUCUCCGCCUGCCUGUAGACUCUUGUAGGCCUGCCGCUGCUCCACG .....................(((((....((((((..(((((((((((((....)))))).))...((.((....)).)).))))).))))))..)))))................. (-26.33 = -26.03 + -0.30)

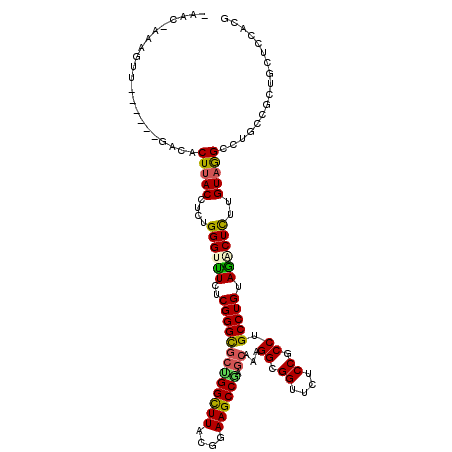
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:02 2006