
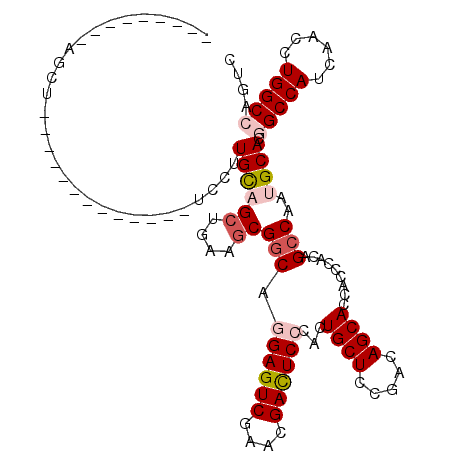
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,793,633 – 5,793,728 |
| Length | 95 |
| Max. P | 0.969450 |

| Location | 5,793,633 – 5,793,728 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.88 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -23.46 |
| Energy contribution | -24.68 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5793633 95 + 20766785 ---------AGCU-------------UCCUUGCAGCUGAAGCGGCAGGAGUCGAACGACUCCCACUGCUCUGACAGCACCACCCACAGCCAAUGCACGGCCAUCAACCUGGCCAGUC ---------.((.-------------.....)).((((.(((((..((((((....))))))..)))))....))))..........((....))..(((((......))))).... ( -33.70) >DroVir_CAF1 124054 108 + 1 ---------AGCAUCCAAUCCUUAACCCCUUGUUGCUCAAUCGACAAGAGUCGAACGAUUCGCACUGCUCGGACAGCACACAGCACAGCCAAUGCACUGCCAUACAUCUGGCCUCGC ---------.((((....(((.......(((((((......)))))))((((((.....))).)))....)))..((.....)).......))))...((((......))))..... ( -21.90) >DroSim_CAF1 85251 95 + 1 ---------AGCU-------------UCCUUGCAGCUGAAGCGGCAGGAGUCUAACGACUCCCACUGCUCCGACAGCACCACCCACAGCCAAUGCACGGCCAUCAACCUGGCCAGUC ---------.((.-------------.....)).((((.(((((..((((((....))))))..)))))....))))..........((....))..(((((......))))).... ( -35.10) >DroEre_CAF1 87125 95 + 1 ---------AGCU-------------UCCUUGCAGCUGAAGCGGCAGGAGUCGAACGACUCCCACUGCUCCGACAGCACCACCCACAGCCAAUGCACGGCCAUCAACCUGGCCAGUC ---------.((.-------------.....)).((((.(((((..((((((....))))))..)))))....))))..........((....))..(((((......))))).... ( -33.70) >DroYak_CAF1 85540 95 + 1 ---------AGCU-------------UCCUUGCAGCUGAAGCGGCAGGAGUCGAACGACUCCCACUGCUCCGACAGCACCACCCACAGCCAAUGCACGGCCAUCAACCUGGCCAGUC ---------.((.-------------.....)).((((.(((((..((((((....))))))..)))))....))))..........((....))..(((((......))))).... ( -33.70) >DroMoj_CAF1 125470 117 + 1 AAUUUGAUUUGCAUGCCAAAGUGAAUCCCUUGUUGCUCUCGCGGCAAGAGUCGAACGAUUCGCACUGCUCGGACAGCACACAGCACAGCCAAUGCACCGCCAUUAACAUGGCCUCAC ....(((..(((((......(((((((.((((((((....)))))))).(.....)))))))).......((...((.....))....)).)))))..(((((....))))).))). ( -31.00) >consensus _________AGCU_____________UCCUUGCAGCUGAAGCGGCAGGAGUCGAACGACUCCCACUGCUCCGACAGCACCACCCACAGCCAAUGCACGGCCAUCAACCUGGCCAGUC ..............................((((((....))(((.((((((....))))))...((((.....)))).........)))..)))).(((((......))))).... (-23.46 = -24.68 + 1.22)


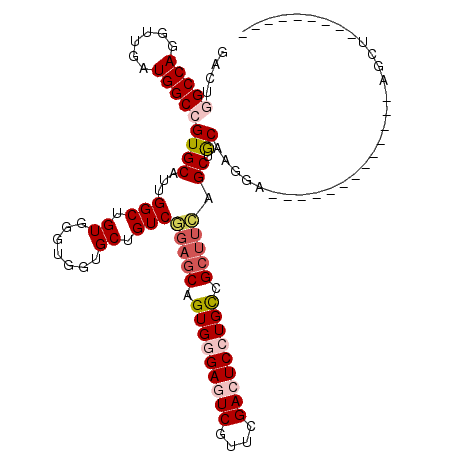
| Location | 5,793,633 – 5,793,728 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.88 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5793633 95 - 20766785 GACUGGCCAGGUUGAUGGCCGUGCAUUGGCUGUGGGUGGUGCUGUCAGAGCAGUGGGAGUCGUUCGACUCCUGCCGCUUCAGCUGCAAGGA-------------AGCU--------- ....(((((......))))).((((...((((.((((((((((((....)))))(((((((....))))))))))))))))))))))....-------------....--------- ( -42.80) >DroVir_CAF1 124054 108 - 1 GCGAGGCCAGAUGUAUGGCAGUGCAUUGGCUGUGCUGUGUGCUGUCCGAGCAGUGCGAAUCGUUCGACUCUUGUCGAUUGAGCAACAAGGGGUUAAGGAUUGGAUGCU--------- (((.((((((.(((((....))))))))))).))).((((.(.((((((((.((....)).))))..(((((((.(......).))))))).....)))).).)))).--------- ( -35.80) >DroSim_CAF1 85251 95 - 1 GACUGGCCAGGUUGAUGGCCGUGCAUUGGCUGUGGGUGGUGCUGUCGGAGCAGUGGGAGUCGUUAGACUCCUGCCGCUUCAGCUGCAAGGA-------------AGCU--------- (((.(((((......)))))..((((((.(....).)))))).)))(((((.(..((((((....))))))..).)))))((((.......-------------))))--------- ( -44.10) >DroEre_CAF1 87125 95 - 1 GACUGGCCAGGUUGAUGGCCGUGCAUUGGCUGUGGGUGGUGCUGUCGGAGCAGUGGGAGUCGUUCGACUCCUGCCGCUUCAGCUGCAAGGA-------------AGCU--------- (((.(((((......)))))..((((((.(....).)))))).)))(((((.(..((((((....))))))..).)))))((((.......-------------))))--------- ( -43.90) >DroYak_CAF1 85540 95 - 1 GACUGGCCAGGUUGAUGGCCGUGCAUUGGCUGUGGGUGGUGCUGUCGGAGCAGUGGGAGUCGUUCGACUCCUGCCGCUUCAGCUGCAAGGA-------------AGCU--------- (((.(((((......)))))..((((((.(....).)))))).)))(((((.(..((((((....))))))..).)))))((((.......-------------))))--------- ( -43.90) >DroMoj_CAF1 125470 117 - 1 GUGAGGCCAUGUUAAUGGCGGUGCAUUGGCUGUGCUGUGUGCUGUCCGAGCAGUGCGAAUCGUUCGACUCUUGCCGCGAGAGCAACAAGGGAUUCACUUUGGCAUGCAAAUCAAAUU .(((.((.(((((((.(((((..(((.(((...))))))..)))))(((((.((....)).))))).((((((..((....))..)))))).......)))))))))...))).... ( -37.50) >consensus GACUGGCCAGGUUGAUGGCCGUGCAUUGGCUGUGGGUGGUGCUGUCGGAGCAGUGGGAGUCGUUCGACUCCUGCCGCUUCAGCUGCAAGGA_____________AGCU_________ ....(((((......)))))((((...(((.((.......)).)))(((((.(((((((((....))))))))).))))).)).))............................... (-28.16 = -29.13 + 0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:00 2006