
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,792,504 – 5,792,609 |
| Length | 105 |
| Max. P | 0.507098 |

| Location | 5,792,504 – 5,792,609 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5792504 105 - 20766785 GCGGCGACUGCAGGG-----AAUUGAAUUCCCCGGAGCUGAAGUCGCUGUUCAGUUCCAUUUCCUGGAUUUGCACAUUGCGCGAGAAGUUCGAAAACUGGCCACUUGACA ..(((...(((((((-----(((...)))))).(((((((((.......)))))))))............))))..(((.((.....)).)))......)))........ ( -29.10) >DroVir_CAF1 121679 102 - 1 CUGGCGACUGCAGCGGCCCA--UUGAACUCACCGGAGCUACAAUCGCUGCUCAGCUCCAGCUCUUGAAUUUGCACAUCGCGCGCGAAUUUACUAAACUG------UUGCA ((((.(.(((.((((((..(--(((..(((....)))...)))).)))))))))).))))....(((((((((.(.....).)))))))))........------..... ( -29.30) >DroSec_CAF1 83749 105 - 1 ACGGCGACUGCAGGG-----AAUUGAAUUCCCCGGAGCUGCAGUCGCUGUUCAGUUCCAUCUCCUGGAUUUGCACAUCGCGCGAGAAGCGCGAAAAUUGGCCAUUUGACA (((((((((((((((-----(((...))))))......))))))))))))((((.((((.....))))...((.((((((((.....))))))....))))...)))).. ( -40.90) >DroEre_CAF1 85978 105 - 1 GCGGCGACUGCAGGG-----AAUUGAAUUCCCCGGAGCUGCAGUCGCUGUUCAGUUCCAGCUCCUGGAUUUGCACGUCGCGCGAGAAGCGCGAGAACUGACCACUUGACA (((((((((((((((-----(((...))))))......))))))))))))(((((((..((..........))...((((((.....))))))))))))).......... ( -46.20) >DroYak_CAF1 83799 105 - 1 GCGGCGACUGCAGGG-----AAUUGAAUUCCCCGGAGCUGAAUUCGCUGUUCAGUUCCAGCUCCUGGAUUUGCACAUCACGCGAAAAGCGCGCAAACUGACCACUUGACA (((((....((.(((-----(((...)))))).((((((((((.....)))))))))).)).......(((((.......)))))..)).)))................. ( -33.50) >DroMoj_CAF1 121879 102 - 1 GCGGCGAUUGCAGCGGCCCA--UUUAACUCACCGGAGCUGCAGUCGCUGCUCUGCUCCAGCUCCUGAAUCUGCAAGUCGCGCGAGAAUUUACUGAACUG------UUGCA ((((((((((((((..((..--...........)).))))))))))))))...((..(((.((.(((((((((.......)).)).)))))..)).)))------..)). ( -38.22) >consensus GCGGCGACUGCAGGG_____AAUUGAAUUCCCCGGAGCUGCAGUCGCUGUUCAGUUCCAGCUCCUGGAUUUGCACAUCGCGCGAGAAGCGCGAAAACUG_CCACUUGACA (((((((((((((..............(((....))))))))))))))))...((((((.....))))(((((.......)))))......................)). (-20.98 = -21.40 + 0.42)

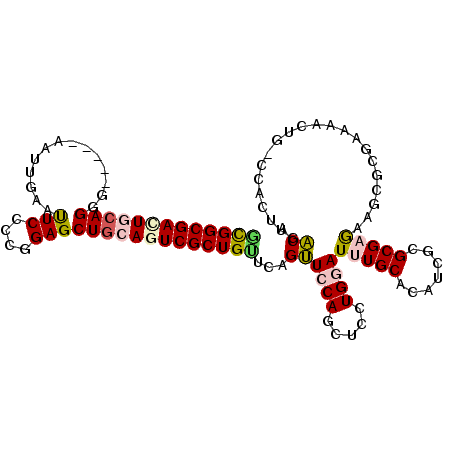
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:58 2006