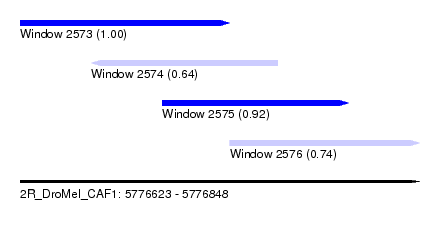
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,776,623 – 5,776,848 |
| Length | 225 |
| Max. P | 0.998224 |

| Location | 5,776,623 – 5,776,741 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -9.66 |
| Energy contribution | -11.74 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.33 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5776623 118 + 20766785 AGUUCUUUAGUCCCAUCUCUUUUAUCCAGAAAUAUGCUCGGUAAAAGAGAAGCCACCGGCGGAUUCUCAGUGAAACUCUCCAAUAAUUAUAGGAGAGUGUAUGUUCUGGCUCUCU--AAC ........(((((..(((((((((((.((.......)).))))))))))).(((...)))))))).((((....(((((((..........))))))).......))))......--... ( -32.10) >DroSec_CAF1 66909 114 + 1 AGUUCUUUAGUGCCAUCUCUUUUAUCCAGAAAUAUGCUCGGUAAAAGAGAAGCCACCGGUGGUUUCUAGGUAAAACUCUCCAAAAAUUAUAGGAGAGUGUAGG----GGCUCUCU--AAC (((((((...((((...(((((((((.((.......)).)))))))))(((((((....)))))))..))))..(((((((..........)))))))..)))----))))....--... ( -36.00) >DroSim_CAF1 68046 114 + 1 AGUUCUUUAGUCCCAUCUCUUUUAUCCAGAAAUAUGCUCGGUAAAAGAGAAGCCACCGGUGGUUUCUCUGUAAAACUCUCCAAAAAUUAUAGGAGAGUGUAGG----GGCUCUCU--AAC ........((((((......((((((.((.......)).))))))((((((((((....)))))))))).....(((((((..........)))))))...))----))))....--... ( -37.10) >DroEre_CAF1 70027 94 + 1 AGUUGUUUAAUCGCAUCUCUUUUAUCCACAAAAGUGUUCGUUAAAAGAGAAUCCACCGGCUAUUGCUCGGUGAAACACUCCAA--------------------------CUCUCCAAAAC (((((..........(((((((((..(((....))).....)))))))))...(((((((....)).)))))........)))--------------------------))......... ( -24.00) >DroYak_CAF1 67519 94 + 1 AGUUGCUUAGUCCCAUCUGUUUUAUGCACUAAUAGGCUCGUUAAAAGAGAAGAAACCGUCGGUUUCUCGGCGAAACUCUCCAA--------------------------CUCUCCAAAAC (((((.....((((.(((.(((((.((........))....))))).)))(((((((...))))))).)).)).......)))--------------------------))......... ( -18.30) >consensus AGUUCUUUAGUCCCAUCUCUUUUAUCCAGAAAUAUGCUCGGUAAAAGAGAAGCCACCGGCGGUUUCUCGGUGAAACUCUCCAA_AAUUAUAGGAGAGUGUA_G____GGCUCUCU__AAC ...............(((((((((((.((.......)).))))))))))).......((.((((((.....))))))..))....................................... ( -9.66 = -11.74 + 2.08)


| Location | 5,776,663 – 5,776,768 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -22.13 |
| Energy contribution | -22.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.642012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5776663 105 - 20766785 CUGU----GUGCAUACAGUGUGUGUUUUACAGUUAGAGAGCCAGAACAUACACUCUCCUAUAAUUAUUGGAGAGUUUCACUGAGAAUCCGCCGGUGGCUUCUCUUUUAC ((((----(.((((((...))))))..)))))..((((((((.(((.....(((((((..........))))))))))((((.(......)))))))).)))))..... ( -31.50) >DroSec_CAF1 66949 101 - 1 CUAU----GUGCAUACAGUGUGUGUUUUACAGUUAGAGAGCC----CCUACACUCUCCUAUAAUUUUUGGAGAGUUUUACCUAGAAACCACCGGUGGCUUCUCUUUUAC ...(----((((((((...))))))...)))...((((((((----((...(((((((..........)))))))........(....)...)).))).)))))..... ( -25.40) >DroSim_CAF1 68086 105 - 1 CUAUGUAUGUGCAUACAGUGUGUGUUUUACAGUUAGAGAGCC----CCUACACUCUCCUAUAAUUUUUGGAGAGUUUUACAGAGAAACCACCGGUGGCUUCUCUUUUAC ...((((...((((((...))))))..))))...((((((((----((...(((((((..........)))))))......(......)...)).))).)))))..... ( -28.80) >consensus CUAU____GUGCAUACAGUGUGUGUUUUACAGUUAGAGAGCC____CCUACACUCUCCUAUAAUUUUUGGAGAGUUUUACAGAGAAACCACCGGUGGCUUCUCUUUUAC (((.......((((((...))))))........)))...............(((((((..........)))))))......(((((.(((....))).)))))...... (-22.13 = -22.03 + -0.11)

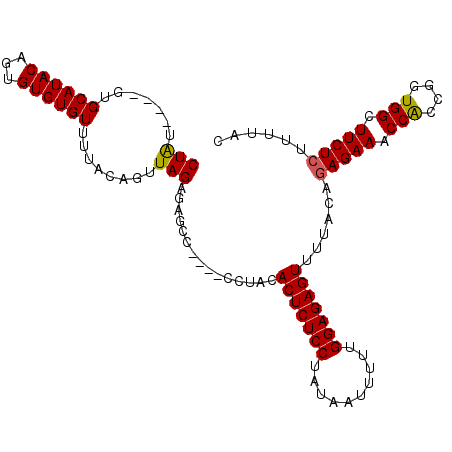
| Location | 5,776,703 – 5,776,808 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5776703 105 + 20766785 CAAUAAUUAUAGGAGAGUGUAUGUUCUGGCUCUCUAACUGUAAAACACACACUGUAUGCAC----ACAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGA ......((((((((((((..........))))))...))))))..........((((((((----....))))))))....((((.((((((......)))))).)))) ( -25.60) >DroSec_CAF1 66989 101 + 1 CAAAAAUUAUAGGAGAGUGUAGG----GGCUCUCUAACUGUAAAACACACACUGUAUGCAC----AUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGA ......((((((((((((.....----.))))))...))))))..........((((((((----....))))))))....((((.((((((......)))))).)))) ( -26.70) >DroSim_CAF1 68126 105 + 1 CAAAAAUUAUAGGAGAGUGUAGG----GGCUCUCUAACUGUAAAACACACACUGUAUGCACAUACAUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGA ......((((((((((((.....----.))))))...))))))..........((((((((........))))))))....((((.((((((......)))))).)))) ( -26.70) >consensus CAAAAAUUAUAGGAGAGUGUAGG____GGCUCUCUAACUGUAAAACACACACUGUAUGCAC____AUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGA ......((((((((((((..........))))))...))))))..........((((((((........))))))))....((((.((((((......)))))).)))) (-24.67 = -24.67 + 0.00)

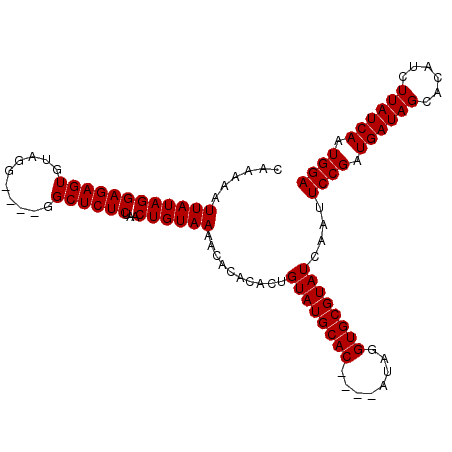

| Location | 5,776,741 – 5,776,848 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5776741 107 + 20766785 UGUAAAACACACACUGUAUGCAC-------------ACAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGAACCGAGCUGAUAAAAGCGGGAACCCAUCGGACUGGCAAAC (((............((((((((-------------....))))))))...(((((.((((((......)))))).)))))(((((((......)))((....)).))))....)))... ( -29.90) >DroSec_CAF1 67023 103 + 1 UGUAAAACACACACUGUAUGCAC-------------AUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGAACCGAGCUGAUAAAAGCGGGAACCCAUCGGGCAGGC---- .............((((((((((-------------....)))))).....(((((.((((((......)))))).)))))(((((((......)))((....)).))))))))..---- ( -31.60) >DroSim_CAF1 68160 107 + 1 UGUAAAACACACACUGUAUGCACAU---------ACAUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGAACCGAGCUGAUAAAAGCGGGAACCCAUCGGGCAGGC---- .............((((((((((..---------......)))))).....(((((.((((((......)))))).)))))(((((((......)))((....)).))))))))..---- ( -31.60) >DroEre_CAF1 70121 115 + 1 UGUAAAACACACAAUGCAUGCACAUGCUGUAUGUACAUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGAACCGAGCUGAUAAAAGCGAGAACUCAACG-GCAGGC---- ......((.(((..((((((((.....)))))))).....))).)).....(((((.((((((......)))))).)))))(((.(((......)))((....))..))-).....---- ( -26.90) >DroYak_CAF1 67613 102 + 1 UGUAAAGCACACAAUGCAUGCAC-------------AUAGGUGCGUAUCAAUUCCGAUGAUAGCGCACCUUAUCAAUGGAACCGAGCUGAUAAAACCGGGAACUCAUCG-GCAGGC---- ((((..(((.....))).))))(-------------((((((((((((((.......)))).)))))))).....)))...((..((((((..............))))-)).)).---- ( -24.64) >consensus UGUAAAACACACACUGUAUGCAC_____________AUAGGUGCGUAUCAAUUCCGAUGAUAGCACAUCUUAUCAAUGGAACCGAGCUGAUAAAAGCGGGAACCCAUCGGGCAGGC____ .............((((((((((.................)))))).....(((((.((((((......)))))).)))))(((((((......)))((....)).))))))))...... (-21.55 = -22.15 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:49 2006