
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,753,463 – 5,753,568 |
| Length | 105 |
| Max. P | 0.872686 |

| Location | 5,753,463 – 5,753,568 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -24.05 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
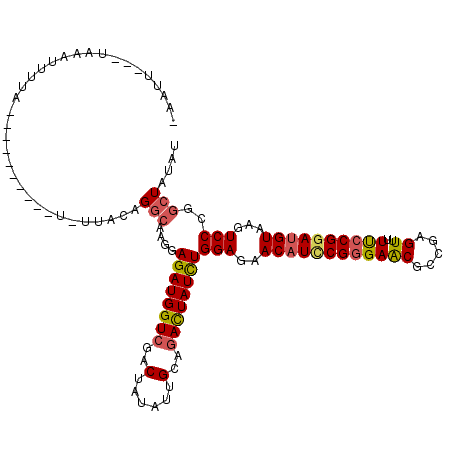
>2R_DroMel_CAF1 5753463 105 + 20766785 -AAAG---UACAUUUUA----------U-UUACAGGCAAGGAGAUGGUCGACUAUAUUGCAGACUAUUUGGAGAACAUCCGGGAACGCCGAGUUUUUCCGGAUGUCAGUCCGGGCUAUAU -....---.........----------.-.....(((..((((((((((..(......)..)))))))....((.((((((((((.((...)).))))))))))))..)))..))).... ( -31.40) >DroSim_CAF1 44833 105 + 1 -AAUG---UAAACUUUA----------U-UUACAGGCAAGGAGAUGGUCGACUAUAUUGCGGACUAUCUGGAGAACAUCCGGGAACGCCGAGUUUUUCCGGAUGUGAGUCCCGGCUACAU -..((---((((.....----------)-)))))(((....((((((((..(......)..))))))))(((..(((((((((((.((...)).)))))))))))...)))..))).... ( -34.70) >DroEre_CAF1 44031 111 + 1 -AAUU---UCCAUUUUA----UUUCCAU-UUACAGGCAAGGAGAUGGUCGACUACAUCGCAGACUAUCUGGAGAACAUCCGGGAGCGGCGAGUUUUUCCGGAUGUAAGUCCCGGAUAUAU -....---(((......----(((((..-...(......).((((((((..(......)..)))))))))))))((((((((((((.....))..)))))))))).......)))..... ( -32.30) >DroYak_CAF1 44082 111 + 1 -AACU---AAAAUUGUA----UUUCCCU-UUACAGGCAAGGAGAUGGUCGACUAUAUUGCAGACUAUCUGGAGAACAUCCGGGAACGCCGAGUUUUUCCGGAUGUAAGUCCCGGCUAUAU -....---.....((((----.......-.))))(((..((((((((((..(......)..)))))))......(((((((((((.((...)).)))))))))))...)))..))).... ( -34.70) >DroMoj_CAF1 69527 119 + 1 AAAUUCAAACAAGUUAAGCUAUUUUUUU-UCGCAGGCAAGGAGAUGGUAGACUAUAUAGCAGACUAUUUGGAGAACAUACGGGAGCGUCGCGUCUUUCCGGAUGUGAGUCCGGGCUAUAU ...........((((..(((((((((((-.(....).))))))))))).))))(((((((.........(((((....(((....)))....)))))((((((....))))))))))))) ( -30.90) >DroAna_CAF1 44783 106 + 1 -AAAA---UAACUAUAA----------UGUUUUAGGAAAAGAGAUGGUCGACUAUAUAGCCGAUUAUUUGGAGAACAUUCGAGAGCGACGCGUCUUCCCGGACGUAAGUCCCGGCUAUAU -....---..(((((..----------..(((....)))....))))).....(((((((((.......(((((....(((....)))....)))))..((((....))))))))))))) ( -27.60) >consensus _AAUU___UAAAUUUUA__________U_UUACAGGCAAGGAGAUGGUCGACUAUAUUGCAGACUAUCUGGAGAACAUCCGGGAACGCCGAGUUUUUCCGGAUGUAAGUCCCGGCUAUAU ..................................(((....((((((((..(......)..))))))))(((..((((((((((((.....))..))))))))))...)))..))).... (-24.05 = -24.17 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:40 2006