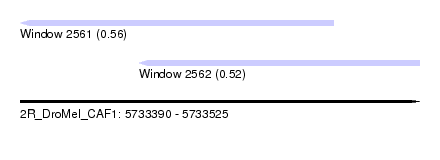
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,733,390 – 5,733,525 |
| Length | 135 |
| Max. P | 0.556371 |

| Location | 5,733,390 – 5,733,496 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Mean single sequence MFE | -16.74 |
| Consensus MFE | -7.63 |
| Energy contribution | -7.38 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5733390 106 - 20766785 CAAAGAUACGAAUUGUGUAUGUAACACA-----U--GUGUCAUAAAAAGCG-UACAGCAGGCGAUCAUAAUAAAAUAAAAUGGAAAAGAAACUUGCGAAUUAAAAUUCGCGCGU ....((((((...(((((.....)))))-----)--))))).......(((-(...(((((...((.....................))..)))))((((....)))))))).. ( -20.50) >DroPse_CAF1 24984 111 - 1 CAAAGAUACAAAUUGUAUAUGUAACAAAAAAUAUGCGAGCAAUAAAGAGAUUCACAAA---AAAGAAUAAUAAAACAAAAUGGAAAAGAAACUUGCGAAUUAAAGCUCGCGCGC ............((((((((..........))))))))((.........((((.....---...))))..........................((((........)))))).. ( -13.60) >DroSec_CAF1 23904 97 - 1 CAAAGAUACAAAUUGUGUAUGUAACACA-----U--GUGUCAU----------ACAGCAGGCGAUCAUAAUAAAAUAAAAUUGAAAAGAAACUUGCGAAUUAAAAUUCGCGCGU ....((((((...(((((.....)))))-----)--)))))..----------...((......(((..............)))..........((((((....)))))))).. ( -18.94) >DroYak_CAF1 23895 104 - 1 --AAGAUACAAAUUGUGUAUGUAACACA-----U--GUGUCAUAAAAAGCG-UACAGAAGGGGAUCAUAAUAAAAUAAAAUGGAAAAGAAACUUGCGAAUUAAAAUUCGCGCGU --..((((((...(((((.....)))))-----)--))))).......(((-(......(((..((.....................))..)))((((((....)))))))))) ( -19.60) >DroAna_CAF1 26511 102 - 1 CAAAGAUACAAAUUGUGUAUGUAACUGA-----G--CUAUAAAAAAAAGUG-UACAAAAAAAGAUCA----AAAAAGAAAUGAAAAAGAAACUUGCGAAUUUCAAUUCGCAGGC ....(((........(((((...(((..-----.--...........))))-)))).......))).----....................(((((((((....))))))))). ( -15.00) >DroPer_CAF1 25173 112 - 1 CAAAGAUACAAAUUGUAUAUGUAACAAAAAAUAUGCGAGCAAUAAAGAGAUUCACAAAA--AAAGAAUAAUAAAACAAAAUGGAAAAGAAACUUGCGAAUUAAAGCUCGCGCGC .....((((.....))))................((((((........(((((.(((..--...............................))).)))))...)))))).... ( -12.81) >consensus CAAAGAUACAAAUUGUGUAUGUAACACA_____U__GUGUAAUAAAAAGAG_UACAAAAGGAGAUCAUAAUAAAAUAAAAUGGAAAAGAAACUUGCGAAUUAAAAUUCGCGCGC .....((((.....))))...........................................................................(((((((....)))))))... ( -7.63 = -7.38 + -0.25)


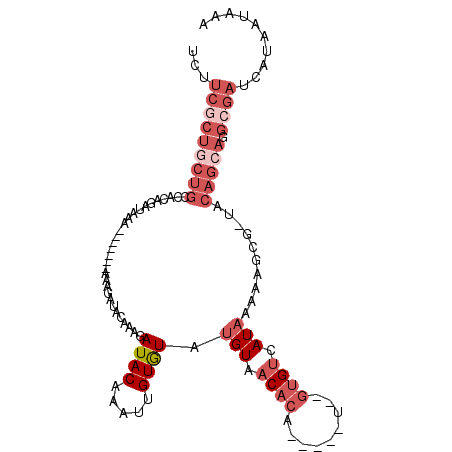
| Location | 5,733,430 – 5,733,525 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -6.16 |
| Energy contribution | -8.52 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.33 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5733430 95 - 20766785 UCUUCGCUGCUGCCACAGAUAAA--------AAGAUACAAAGAUACGAAUUGUGUAUGUAACACA-----U--GUGUCAUAAAAAGCG-UACAGCAGGCGAUCAUAAUAAA ...(((((((((..((..((...--------...)).....((((((...(((((.....)))))-----)--))))).........)-).))))).)))).......... ( -20.10) >DroSec_CAF1 23944 94 - 1 UCUUCGCUGCUGCCACAGAUAAAUAGAUACAAAGAUACAAAGAUACAAAUUGUGUAUGUAACACA-----U--GUGUCAU----------ACAGCAGGCGAUCAUAAUAAA ...(((((((((......((......)).............((((((...(((((.....)))))-----)--)))))..----------.))))).)))).......... ( -20.00) >DroSim_CAF1 23125 103 - 1 UCUUCGCUGCUGCCACAGAUAAAUAGAUACAAAGAUACAAAGAUACAAAUUGUGUAUGUAACACA-----U--GUGUCAUAAAAAGCG-UACAGCAGGCGAUCAUAAUAAA ...(((((((((..((.........((((((..(.((((...((((.....)))).)))).)...-----)--))))).........)-).))))).)))).......... ( -21.47) >DroEre_CAF1 23910 96 - 1 UCUUCGCAGCUGCCGCAGAUAAA-------AAAGAUACAAAGAUACAAAUUGUGUAUGUAACACA-----U--GUGUCAUAAAAAGCG-UACAGCAGGCGAUCAUAAUAAA ...((((.((((.(((..((...-------....)).....((((((...(((((.....)))))-----)--))))).......)))-..))))..)))).......... ( -21.80) >DroYak_CAF1 23935 87 - 1 UCUUCGCUGCUGCCUCAGAUAAA----------------AAGAUACAAAUUGUGUAUGUAACACA-----U--GUGUCAUAAAAAGCG-UACAGAAGGGGAUCAUAAUAAA (((.((((.(((...))).....----------------..((((((...(((((.....)))))-----)--)))))......))))-...)))................ ( -16.00) >DroPer_CAF1 25213 90 - 1 UC---GCUC--------GACC--------AAAAUAUACAAAGAUACAAAUUGUAUAUGUAACAAAAAAUAUGCGAGCAAUAAAGAGAUUCACAAAA--AAAGAAUAAUAAA ..---((((--------(...--------...((((((((.........))))))))...............))))).........((((......--...))))...... ( -11.50) >consensus UCUUCGCUGCUGCCACAGAUAAA_______AAAGAUACAAAGAUACAAAUUGUGUAUGUAACACA_____U__GUGUCAUAAAAAGCG_UACAGCAGGCGAUCAUAAUAAA ...(((((((((..............................((((.....)))).(((.((((.........)))).)))..........))))).)))).......... ( -6.16 = -8.52 + 2.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:36 2006