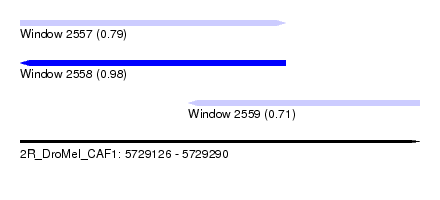
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,729,126 – 5,729,290 |
| Length | 164 |
| Max. P | 0.982467 |

| Location | 5,729,126 – 5,729,235 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5729126 109 + 20766785 CACGCGGGCGCAAGUAAUCACGAAUGUAGUAUACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUAG ...(((.((....))..((((((((((((((((..........)))))......)))))))))))........))).............((((((((....)))))))) ( -27.70) >DroSec_CAF1 19539 106 + 1 CAGGCGGGCGCAAGUAAUCACGAAUGU---AUACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUAG ..((((.(((..(((..((((((((((---((.(.................).))))))))))))...)))..)))......))))...((((((((....)))))))) ( -27.83) >DroEre_CAF1 19728 103 + 1 ---GCGCUCGCAAGUAAUCACGAAUGU---AUACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUAG ---(((..(....)...((((((((((---((.(.................).))))))))))))........))).............((((((((....)))))))) ( -23.83) >DroYak_CAF1 19585 104 + 1 --CACGCGCAGAAGUAAUCACGAAUGU---AUACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUAG --...(((..((.((..((((((((((---((.(.................).))))))))))))...)))).))).............((((((((....)))))))) ( -24.33) >consensus __CGCGCGCGCAAGUAAUCACGAAUGU___AUACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUAG .....(.(((..(((..((((((((((..........((....))..........))))))))))...)))..))).)...........((((((((....)))))))) (-20.65 = -21.15 + 0.50)


| Location | 5,729,126 – 5,729,235 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -31.31 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5729126 109 - 20766785 CUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUAUACUACAUUCGUGAUUACUUGCGCCCGCGUG ((((((........))))))....((((...(.(((..(((..(((((((((.(((((.((((....))))....)))))....))))))))).)))..)))).)))). ( -35.70) >DroSec_CAF1 19539 106 - 1 CUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUAU---ACAUUCGUGAUUACUUGCGCCCGCCUG ((((((........))))))...........(.(((..(((..(((((((((.(((((.((((....))))....))))---).))))))))).)))..))).)..... ( -36.20) >DroEre_CAF1 19728 103 - 1 CUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUAU---ACAUUCGUGAUUACUUGCGAGCGC--- ((((((........))))))...........((.((..(((..(((((((((.(((((.((((....))))....))))---).))))))))).)))..)).))..--- ( -34.70) >DroYak_CAF1 19585 104 - 1 CUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUAU---ACAUUCGUGAUUACUUCUGCGCGUG-- ((((((........))))))...........(((((.((((..(((((((((.(((((.((((....))))....))))---).))))))))).)))).)))))...-- ( -35.30) >consensus CUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUAU___ACAUUCGUGAUUACUUGCGCCCGCG__ ((((((........))))))...........(((((..(((..(((((((((..((((.((((....))))....)))).....))))))))).)))..)))))..... (-31.31 = -31.88 + 0.56)


| Location | 5,729,195 – 5,729,290 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -18.34 |
| Consensus MFE | -13.80 |
| Energy contribution | -13.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5729195 95 - 20766785 CAAGCGCCCC-------CCUUCCACCCCACAGUC--CCAAUCCC------C-AACCAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAG ..........-------.................--........------.-.......((((((.......)))))).(((..(((((((.........))))))).))) ( -13.80) >DroSec_CAF1 19605 81 - 1 ---------C-------CCUUCCUCCCCUCAAU-------UCCC------C-UAACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAG ---------.-------..........((((..-------....------.-....((.((((((.......)))))).))...(((((((.........))))))))))) ( -14.00) >DroEre_CAF1 19791 104 - 1 GG-AGACGCCCCGUCCCCCUCCCUCCGCCCCUUCCACCAUUCCC------CACCACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAG ((-.((((...)))).))..........................------((((((((..((((.......((((((........))))))....)))).))))).))).. ( -23.40) >DroYak_CAF1 19649 108 - 1 AG-GAGCCCCCCGUCCCCUUCCC--CACCUCUUCCGCCGCUCCCUCAUUCCCCCACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAG .(-((((................--.............)))))(((((.....(((((..((((.......((((((........))))))....)))).))))).))))) ( -22.15) >consensus _G_G_GCCCC_______CCUCCCUCCCCCCAUUC__CCA_UCCC______C_CAACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAG ...........................................................((((((.......)))))).(((..(((((((.........))))))).))) (-13.80 = -13.80 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:33 2006