
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,710,185 – 5,710,288 |
| Length | 103 |
| Max. P | 0.500000 |

| Location | 5,710,185 – 5,710,288 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -26.93 |
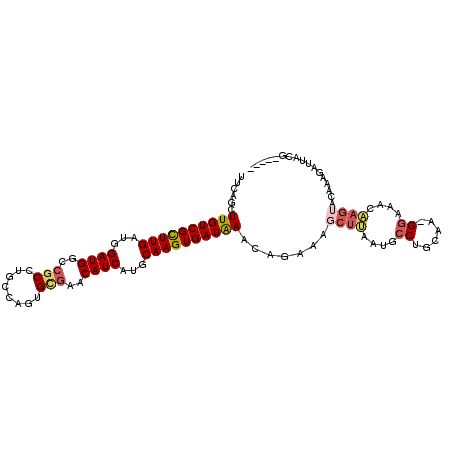
| Consensus MFE | -20.00 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
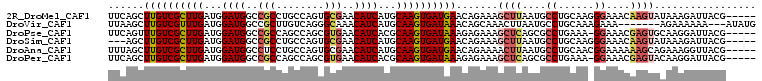
>2R_DroMel_CAF1 5710185 103 + 20766785 UUCAGCUUGUCGCUUGAUGGAUGGCCGCCUGCCAGUGCGAACAUCAUGCAAGUGAUGAACAGAAAGCUUAAUGCCUGCAAGGGAAACAAGUAUAAAGAUUACG----- (((.(.(..(((((((...((((..((((.....).)))..))))...)))))))..).).))).((((....(((....)))....))))............----- ( -28.00) >DroVir_CAF1 6723 98 + 1 UUAAGCUUGUCGUUUGAUGGAUGGCCGCUUGUCAGGGCAAACAUCAUGCAAGUGAUAAACAGCAAACUUAAUGCCUGCAAAGAAA-------AGAAAAAA---AUAUG ....((((((....(((((..((.((........)).))..))))).))))))........(((..(.....)..))).......-------........---..... ( -20.40) >DroPse_CAF1 333 102 + 1 UUCAGUUUGUCGCUUGAUGGAUGGCCGCCAGCCAGCGUGAACAUCACGCAAGUGAUAAAGAGAAAGCUCAGCGCCUGAAA-GGAAACGAGUGCAAGGAUUACG----- (((((((((((((((......((((.....))))(((((.....)))))))))))))))(((....))).....))))).-(....)................----- ( -31.10) >DroSim_CAF1 2 100 + 1 ---AGCUUGUCGCUUGAUGGAUGGCCGCCUGCCAGUGCGAACAUCAUGCAAGUGAUGAACAGAAAGCUUAAUGCCUGCAAGGGAAACAAGUAUAAAGAUUACG----- ---.(.(..(((((((...((((..((((.....).)))..))))...)))))))..).).....((((....(((....)))....))))............----- ( -27.40) >DroAna_CAF1 2232 103 + 1 UUUAGCUUGUCGCUUGAUGGAUGGCCUCCUGCCAGUGCGAACAUCAUGCAAGUGAUGAACAGAAAACUUAAUGCCUGCAACGGAAAAAAGCAGAAAGGUUACG----- ..((((((((((((((((((.((((.....))))((....)).)))).))))))))..................((((...........))))..))))))..----- ( -24.10) >DroPer_CAF1 336 102 + 1 UUCAGCUUGUCGCUUGAUGGAUGGCCGCCAGCCAGCGUGAACAUCACGCAAGUGAUAAAGAGAAAGCUCAGCGCCUGAAA-GGAAACGAGUACAAGGAUUACG----- .....(((((.(((((.....((((.....))))((((....(((((....)))))...(((....))).))))((....-))...)))))))))).......----- ( -30.60) >consensus UUCAGCUUGUCGCUUGAUGGAUGGCCGCCUGCCAGUGCGAACAUCAUGCAAGUGAUAAACAGAAAGCUUAAUGCCUGCAA_GGAAACAAGUACAAAGAUUACG_____ ......((((((((((...((((..(((........)))..))))...)))))))))).......((((....((......))....))))................. (-20.00 = -20.45 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:28 2006