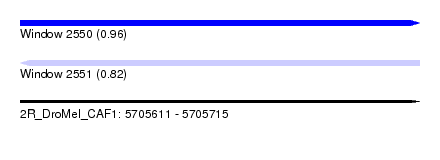
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,705,611 – 5,705,715 |
| Length | 104 |
| Max. P | 0.963297 |

| Location | 5,705,611 – 5,705,715 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -22.73 |
| Energy contribution | -21.90 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
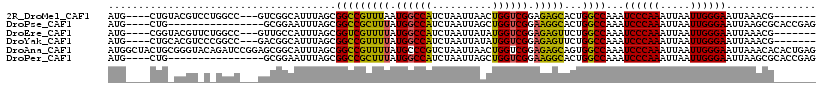
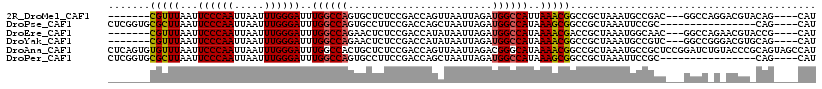
>2R_DroMel_CAF1 5705611 104 + 20766785 AUG----CUGUACGUCCUGGCC---GUCGGCAUUUAGCGGCCGUUUAAUGGCCAUCUAAUUAACUGGUCGGAGAGCACUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAACG------- (((----(((.(((.......)---))))))))((((.((((((...))))))..)))).....(((((((......)))))))..((((((.....))))))........------- ( -33.30) >DroPse_CAF1 30197 98 + 1 AUG----CUG----------------GCGGAAUUUAGCGGCCGCUUUAUGGCCAUCUAAUUAGCUGGUCGGAAGGCACUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAGCGCACCGAG ...----...----------------.(((......(((((((((((.((((((.((....)).)))))).)))))...))))...((((((.....)))))).....))...))).. ( -33.00) >DroEre_CAF1 33700 104 + 1 AUG----CGGUACGUUCUGGCC---GUUGCCAUUUAGCGGUCGUUUUAUGGCCAUCUAAUUAUAUGGUCGGAGAGUUCUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAACG------- ..(----((((........)))---)).(((((..(((....)))..)))))............(((((((......)))))))..((((((.....))))))........------- ( -32.10) >DroYak_CAF1 34300 104 + 1 AUG----CUGCACGUCCCGGCC---GACGGCAUUUAGCGGCCGUUUUAUGGCCAUCUAAUUAUAUGGUCGGAGAGUUCUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAACG------- ..(----((((.((((......---))))))....)))((((((...))))))...........(((((((......)))))))..((((((.....))))))........------- ( -34.60) >DroAna_CAF1 35869 118 + 1 AUGGCUACUGCGGGUACAGAUCCGGAGCGGCAUUUAGCGGCCGUUUUAUGCCCGUCUAAUUAACUGGUCGGAGAGCAGUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAACACACUGAG .((((((((((((((....))))((((((((........))))))))....(((.(((......))).)))...))))))))))..((((((.....))))))............... ( -46.00) >DroPer_CAF1 29928 98 + 1 AUG----CUG----------------GCGGAAUUUAGCGGCCGCUUUAUGGCCAUCUAAUUAGCUGGUCGGAAGGCACUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAGCGCACCGAG ...----...----------------.(((......(((((((((((.((((((.((....)).)))))).)))))...))))...((((((.....)))))).....))...))).. ( -33.00) >consensus AUG____CUG_ACGU_C_GGCC___GGCGGCAUUUAGCGGCCGUUUUAUGGCCAUCUAAUUAACUGGUCGGAGAGCACUGGCCAAAUCCCAAAUUAAUUGGGAAUUAAACG_______ ......................................(((((((((.((((((..........)))))).)))))...))))...((((((.....))))))............... (-22.73 = -21.90 + -0.83)
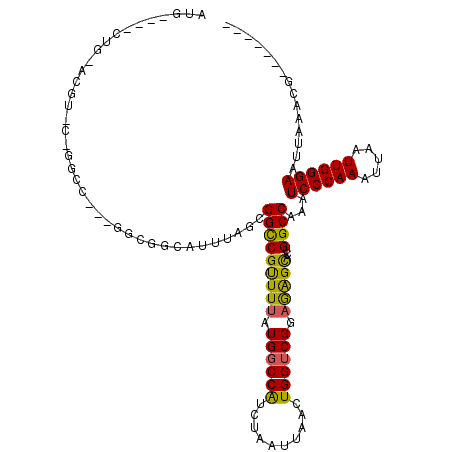
| Location | 5,705,611 – 5,705,715 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
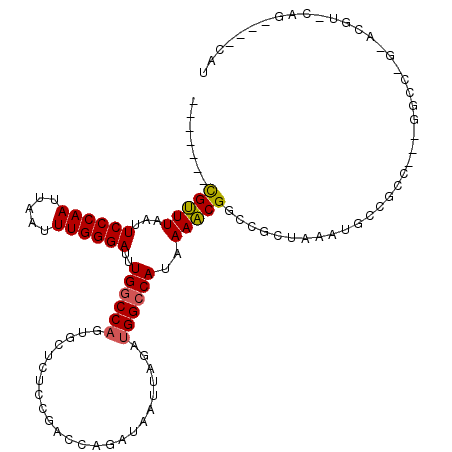
>2R_DroMel_CAF1 5705611 104 - 20766785 -------CGUUUAAUUCCCAAUUAAUUUGGGAUUUGGCCAGUGCUCUCCGACCAGUUAAUUAGAUGGCCAUUAAACGGCCGCUAAAUGCCGAC---GGCCAGGACGUACAG----CAU -------.((((((.((((((.....))))))..((((((...((....((....))....)).))))))))))))((((((........).)---))))...........----... ( -30.50) >DroPse_CAF1 30197 98 - 1 CUCGGUGCGCUUAAUUCCCAAUUAAUUUGGGAUUUGGCCAGUGCCUUCCGACCAGCUAAUUAGAUGGCCAUAAAGCGGCCGCUAAAUUCCGC----------------CAG----CAU .((((.((((((((.((((((.....)))))).)))...)))))...))))...(((........((((.......))))((........))----------------.))----).. ( -30.50) >DroEre_CAF1 33700 104 - 1 -------CGUUUAAUUCCCAAUUAAUUUGGGAUUUGGCCAGAACUCUCCGACCAUAUAAUUAGAUGGCCAUAAAACGACCGCUAAAUGGCAAC---GGCCAGAACGUACCG----CAU -------(((((...((((((.....))))))..(((((..........(.((((........)))).)...........(((....)))...---)))))))))).....----... ( -26.60) >DroYak_CAF1 34300 104 - 1 -------CGUUUAAUUCCCAAUUAAUUUGGGAUUUGGCCAGAACUCUCCGACCAUAUAAUUAGAUGGCCAUAAAACGGCCGCUAAAUGCCGUC---GGCCGGGACGUGCAG----CAU -------(((((...((((((.....))))))..(((((((....))..(((..(((..((((.(((((.......))))))))))))..)))---)))))))))).....----... ( -31.50) >DroAna_CAF1 35869 118 - 1 CUCAGUGUGUUUAAUUCCCAAUUAAUUUGGGAUUUGGCCACUGCUCUCCGACCAGUUAAUUAGACGGGCAUAAAACGGCCGCUAAAUGCCGCUCCGGAUCUGUACCCGCAGUAGCCAU ...............((((((.....))))))..((((.(((((..........(((.....)))(((.(((...(((..((........)).)))....))).)))))))).)))). ( -33.10) >DroPer_CAF1 29928 98 - 1 CUCGGUGCGCUUAAUUCCCAAUUAAUUUGGGAUUUGGCCAGUGCCUUCCGACCAGCUAAUUAGAUGGCCAUAAAGCGGCCGCUAAAUUCCGC----------------CAG----CAU .((((.((((((((.((((((.....)))))).)))...)))))...))))...(((........((((.......))))((........))----------------.))----).. ( -30.50) >consensus _______CGUUUAAUUCCCAAUUAAUUUGGGAUUUGGCCAGUGCUCUCCGACCAGAUAAUUAGAUGGCCAUAAAACGGCCGCUAAAUGCCGCC___GGCC_G_ACGU_CAG____CAU .......(((((...((((((.....))))))..((((((........................))))))..)))))......................................... (-18.67 = -18.43 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:25 2006