| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,698,105 – 5,698,204 |
| Length | 99 |
| Max. P | 0.808634 |

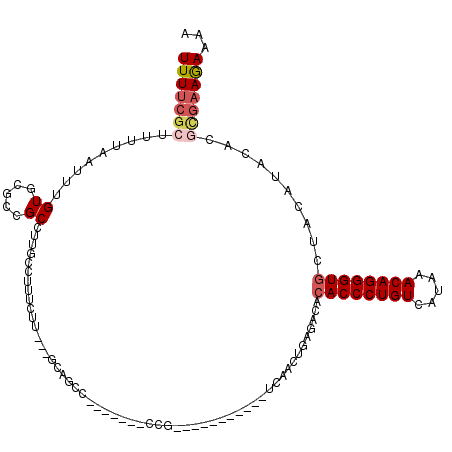
| Location | 5,698,105 – 5,698,204 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5698105 99 + 20766785 UUGUCUUUGCGUGUAUAUAGCACCCUGUUUAUGACAGGGUGUGUCUCAGCUGA-----------CGG-------GGCUGC---AAGAAAGGCAAGGCGCCGCACAAAUUAAAAGCGAAAA ((((((((.(.((((....((((((((((...))))))))))(((((......-----------.))-------))))))---).))))))))).....(((...........))).... ( -31.80) >DroSim_CAF1 28485 99 + 1 UUUUCUUCACGUGUACGUAGCACCCUGUUUAAGACAGGGUGUGUCUCAGUUGA-----------CGG-------CGCUGC---AAGAAAGUCAAGGCGCCGCACAAAUUAAAAGCGAAAA ..........((((.....((((((((((...))))))))))(((......))-----------)((-------((((((---......))...))))))))))................ ( -33.60) >DroEre_CAF1 26492 99 + 1 UUUUCUUCGCGUGUAUGUAGCACCCUGUUUAUGACAGGGUGUGUCUUAGUUGG-----------CGG-------GACUGU---AAGAAAGGCAAAGCGGCGCACAAAUUAAAAGCGAAAA .....(((((((((.(((.((((((((((...))))))))))(((((.....(-----------((.-------...)))---....)))))...)))..)))).........))))).. ( -29.50) >DroYak_CAF1 26978 99 + 1 UUUUUUUCGUAUGUAUGUAGCACCCUGUUUAUGACAGGGUGUGUCUCAGUUGA-----------CGG-------GCCUGC---AAGAAGGGCAAAGCGGCGCACAAAUUAAAAGCGAAAA ...(((((((.....((..((((((((((...))))))))))....))((((.-----------...-------((((..---.....))))....)))).............))))))) ( -31.20) >DroAna_CAF1 28096 119 + 1 -UAUUUGCCAGUGUAAUCAGUACCCUGUUUAUGACAGGGUGUGUCUCAGCUGAGUCCCAAUGGGCGGCGACAUGGGCGGCGAAAAGGGGGGCAUUGCAGCACACAAAUUAAAAGCGAAAA -..(((((.............((((((((...))))))))((((....((((.(((((..(...((.((.......)).))...)..)))))....)))))))).........))))).. ( -36.80) >consensus UUUUCUUCGCGUGUAUGUAGCACCCUGUUUAUGACAGGGUGUGUCUCAGUUGA___________CGG_______GGCUGC___AAGAAAGGCAAAGCGGCGCACAAAUUAAAAGCGAAAA ..........((((.....((((((((((...))))))))))(((((((((.......................))))).........))))........))))................ (-18.90 = -18.90 + 0.00)

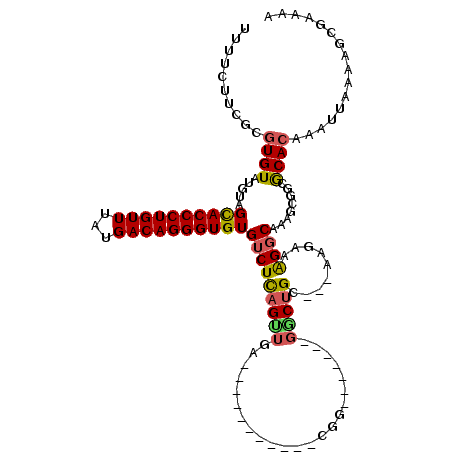
| Location | 5,698,105 – 5,698,204 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.66 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5698105 99 - 20766785 UUUUCGCUUUUAAUUUGUGCGGCGCCUUGCCUUUCUU---GCAGCC-------CCG-----------UCAGCUGAGACACACCCUGUCAUAAACAGGGUGCUAUAUACACGCAAAGACAA .............((((((.((.((..(((.......---))))).-------))(-----------((......))).((((((((.....)))))))).........))))))..... ( -25.10) >DroSim_CAF1 28485 99 - 1 UUUUCGCUUUUAAUUUGUGCGGCGCCUUGACUUUCUU---GCAGCG-------CCG-----------UCAACUGAGACACACCCUGUCUUAAACAGGGUGCUACGUACACGUGAAGAAAA (((((((........((((((((((..((.(......---))))))-------))(-----------((......))).((((((((.....))))))))....))))).)))))))... ( -33.10) >DroEre_CAF1 26492 99 - 1 UUUUCGCUUUUAAUUUGUGCGCCGCUUUGCCUUUCUU---ACAGUC-------CCG-----------CCAACUAAGACACACCCUGUCAUAAACAGGGUGCUACAUACACGCGAAGAAAA ((((((((((((..(((.(((..(((.((.......)---).))).-------.))-----------)))).)))))..((((((((.....))))))))..........)))))))... ( -26.10) >DroYak_CAF1 26978 99 - 1 UUUUCGCUUUUAAUUUGUGCGCCGCUUUGCCCUUCUU---GCAGGC-------CCG-----------UCAACUGAGACACACCCUGUCAUAAACAGGGUGCUACAUACAUACGAAAAAAA ((((((............(.(((((............---)).)))-------.)(-----------((......))).((((((((.....))))))))...........))))))... ( -24.40) >DroAna_CAF1 28096 119 - 1 UUUUCGCUUUUAAUUUGUGUGCUGCAAUGCCCCCCUUUUCGCCGCCCAUGUCGCCGCCCAUUGGGACUCAGCUGAGACACACCCUGUCAUAAACAGGGUACUGAUUACACUGGCAAAUA- .....(((..(((((.((((((......))..(((.....((.((.......)).)).....))).(((....)))))))(((((((.....)))))))...)))))....))).....- ( -28.40) >consensus UUUUCGCUUUUAAUUUGUGCGCCGCCUUGCCUUUCUU___GCAGCC_______CCG___________UCAACUGAGACACACCCUGUCAUAAACAGGGUGCUACAUACACGCGAAGAAAA (((((((.........((.....))......................................................((((((((.....))))))))..........)))))))... (-13.82 = -14.66 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:20 2006