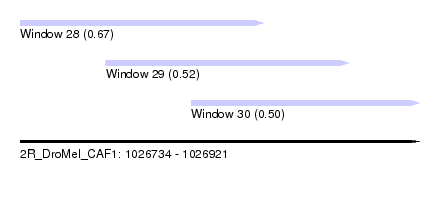
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,026,734 – 1,026,921 |
| Length | 187 |
| Max. P | 0.671220 |

| Location | 1,026,734 – 1,026,848 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -23.04 |
| Energy contribution | -24.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
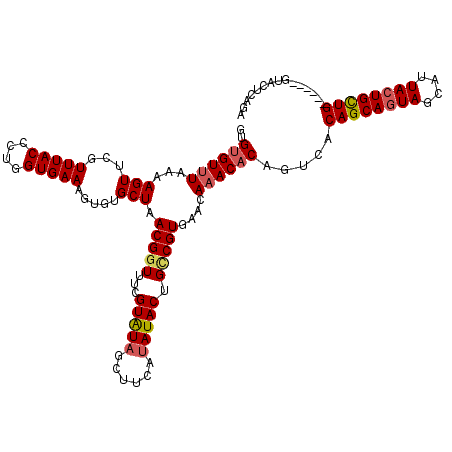
>2R_DroMel_CAF1 1026734 114 + 20766785 GUGUGUUUAAAAGUUCGUUUACCCUGGUGAAAGUGUGCUAACGGUUUCGUAUAGCUCCAUAUACUGUCGUGAACAAACACAGUCACAGCAGUAGCAUUACUGCUG------GCCCUCAGA .(((((((....((((((((((....)))))((((((((((((....))).)))).....)))))....))))))))))))....((((((((....))))))))------......... ( -30.70) >DroSec_CAF1 29163 120 + 1 GUGUGUUUAAAAGUUCGUUUACCCUGGUGAAAGUGUGCUAACGGUUUCGUAUAGCUUUAUAUACUGACGUGAACACACACAGUCACAGCAGUAGCAUUACUGUUGUUUCUGUUACUCAGA (((((((((...(((..(((((....)))))((((((((((((....))).)))).....)))))))).)))))))))((((..(((((((((....)))))))))..))))........ ( -37.70) >DroSim_CAF1 10654 119 + 1 GUGUGUUUA-AAGUUCGUUUACCCUGGUGAAAGUGUGCUAACGGUUUCGUGUAGCUUUAUAUACUGCCGUGAACAAACACAGACGCAGCAGUAGCAUUACUGUUGUUUCUGUUACUCAGA (.(((....-..(((.((((((...(((((((.(((....))).))))(((((......))))).))))))))).)))(((((.(((((((((....))))))))).)))))))).)... ( -36.20) >DroEre_CAF1 86875 114 + 1 GGGAGUUUAAAAGUUCGUUUACUCUGGUGAAAGUGUGCUAACGGUUUUGUAUGCCUUCAAAUACUGCCGUGACGAAACACAGUCUCAGCAAUAGCAAUACUGCUG------GGCCUCAGA .(((((..............)))))(((((..((((....(((((...((((........)))).)))))......))))..))((((((.((....)).)))))------))))..... ( -29.94) >consensus GUGUGUUUAAAAGUUCGUUUACCCUGGUGAAAGUGUGCUAACGGUUUCGUAUAGCUUCAUAUACUGCCGUGAACAAACACAGUCACAGCAGUAGCAUUACUGCUG______GUACUCAGA ..((((((...(((...(((((....))))).....))).(((((...(((((......))))).)))))....)))))).....((((((((....))))))))............... (-23.04 = -24.10 + 1.06)

| Location | 1,026,774 – 1,026,888 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -18.62 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1026774 114 + 20766785 ACGGUUUCGUAUAGCUCCAUAUACUGUCGUGAACAAACACAGUCACAGCAGUAGCAUUACUGCUG------GCCCUCAGAAAGAUCAUCUCACUGCAAUGCAUAUGCAUAGAGUAUUAAA (((....)))...((((((((((((((.((......)))))))..((((((((....))))))))------((...(((..((.....))..)))....)))))))....))))...... ( -25.10) >DroSec_CAF1 29203 120 + 1 ACGGUUUCGUAUAGCUUUAUAUACUGACGUGAACACACACAGUCACAGCAGUAGCAUUACUGUUGUUUCUGUUACUCAGAAAGAUUAUCUCACUGCAAUGCAUAUGCAUAGGGUAUUAAA ...(((..(((((......))))).)))(((....)))((((..(((((((((....)))))))))..))))(((((((..((.....))..))...((((....)))).)))))..... ( -28.10) >DroSim_CAF1 10693 120 + 1 ACGGUUUCGUGUAGCUUUAUAUACUGCCGUGAACAAACACAGACGCAGCAGUAGCAUUACUGUUGUUUCUGUUACUCAGAAAGAUUAUCUCACUGCAAUGCAUAUGCAUAGGGUAUUAAA (((((...(((((......))))).)))))........(((((.(((((((((....))))))))).)))))(((((((..((.....))..))...((((....)))).)))))..... ( -33.60) >DroEre_CAF1 86915 113 + 1 ACGGUUUUGUAUGCCUUCAAAUACUGCCGUGACGAAACACAGUCUCAGCAAUAGCAAUACUGCUG------GGCCUCAGAAAACUUAUCUCACUGCAAGUCAUAUGCAUAGGGUAUUA-A (((((...((((........)))).)))))..........((.(((((((.((....)).)))))------)).)).........(((((...((((.......))))..)))))...-. ( -25.40) >consensus ACGGUUUCGUAUAGCUUCAUAUACUGCCGUGAACAAACACAGUCACAGCAGUAGCAUUACUGCUG______GUACUCAGAAAGAUUAUCUCACUGCAAUGCAUAUGCAUAGGGUAUUAAA (((((...(((((......))))).)))))...............((((((((....)))))))).......................(((..((((.......))))..)))....... (-18.62 = -19.00 + 0.38)

| Location | 1,026,814 – 1,026,921 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
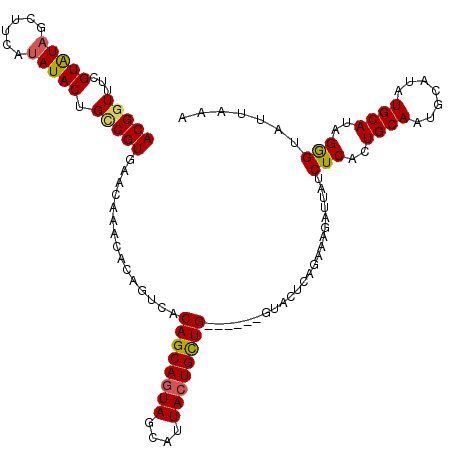
>2R_DroMel_CAF1 1026814 107 + 20766785 AGUCACAGCAGUAGCAUUACUGCUG------GCCCUCAGAAAGAUCAUCUCACUGCAAUGCAUAUGCAUAGAGUAUUAAAGUGCAAAAAAAUA-------AAAUUAACAUGAUACUUUAA .....((((((((....))))))))------........(((((((((.........((((....))))...((((....)))).........-------........))))).)))).. ( -21.50) >DroSec_CAF1 29243 120 + 1 AGUCACAGCAGUAGCAUUACUGUUGUUUCUGUUACUCAGAAAGAUUAUCUCACUGCAAUGCAUAUGCAUAGGGUAUUAAAGUGCAACAAAAUAAAUACAAAAAUUAACGUGAUACUUUAA .(((((.(((((((((....)))).((((((.....)))))).........)))))..(((((((((.....))))....))))).......................)))))....... ( -24.10) >DroSim_CAF1 10733 120 + 1 AGACGCAGCAGUAGCAUUACUGUUGUUUCUGUUACUCAGAAAGAUUAUCUCACUGCAAUGCAUAUGCAUAGGGUAUUAAAGUGCAACAAAAUAAAUACAAAAAUUAACGUGAUACUUUAA (((.(((((((((....))))))))).)))..(((((((..((.....))..))...((((....)))).)))))((((((((..((.....................))..)))))))) ( -24.10) >DroEre_CAF1 86955 113 + 1 AGUCUCAGCAAUAGCAAUACUGCUG------GGCCUCAGAAAACUUAUCUCACUGCAAGUCAUAUGCAUAGGGUAUUA-AGUGCAACACAAUAUAUUUAAAAAUUAACAUGCGGCUUUAA .....(((((.((....)).)))))------((((.......(((((.(((..((((.......))))..)))...))-)))(((........................))))))).... ( -21.76) >consensus AGUCACAGCAGUAGCAUUACUGCUG______GUACUCAGAAAGAUUAUCUCACUGCAAUGCAUAUGCAUAGGGUAUUAAAGUGCAACAAAAUAAAUACAAAAAUUAACAUGAUACUUUAA .....((((((((....))))))))............................((((.......))))((((((((((..((........................)).)))))))))). (-14.34 = -14.84 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:29 2006