
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,670,711 – 5,670,836 |
| Length | 125 |
| Max. P | 0.982975 |

| Location | 5,670,711 – 5,670,801 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.04 |
| Mean single sequence MFE | -20.11 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.28 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5670711 90 + 20766785 CAAAAAAAAAAAGAAACCCUGAGGCGUUCUUCUCACGGCCCCCUAUUACUAGGGUUGAAACAGCCAGCAAAC----AUAU----AUCAA-----UGUGAUUAU ................((.(((((......))))).))..(((((....))))).................(----((((----....)-----))))..... ( -13.70) >DroSec_CAF1 8129 84 + 1 -----AAAAAAGGAUACCCUGGGGCGUUCUUCUCACGGCCCCCUAUUACUAGGGUG-----UGCCGGCAUAC----AUAUGUAUAGGAA-----UGUGAUUAU -----.....(((....)))...((((((((....((((((((((....))))).)-----.))))..((((----....)))))))))-----)))...... ( -24.80) >DroSim_CAF1 8143 82 + 1 -----A--AAAGGAUACCCUGGGGCGUUCUUCUCACGGCCCCCUAUUACUAGGGUG-----UGCCAGCAUAC----AUAUGUAUAGGAA-----UGUGAUUAU -----.--.(((((..((....))..))))).(((((((((((((....))))).)-----.)))...((((----....)))).....-----.)))).... ( -24.50) >DroYak_CAF1 8217 95 + 1 CA---AAAAAAAAAAACCCUGGGGCGUUCUACUCACGGCCCCCUACUACAAGGGUU-----UGUAGACGUAUUAAUACAUUUAAAAAAAUACAAUAAAAUCAU ..---........((((((((((((((.......)).)))))........))))))-----)......(((((..............)))))........... ( -17.44) >consensus _____AAAAAAAGAAACCCUGGGGCGUUCUUCUCACGGCCCCCUAUUACUAGGGUG_____UGCCAGCAUAC____AUAUGUAUAGGAA_____UGUGAUUAU ..............(((((((((((((.......)).)))))........))))))............................................... (-13.52 = -13.28 + -0.25)

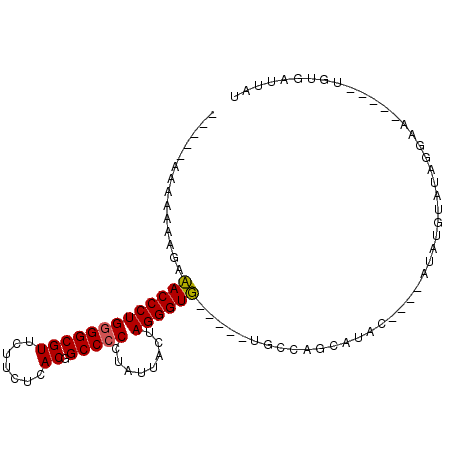
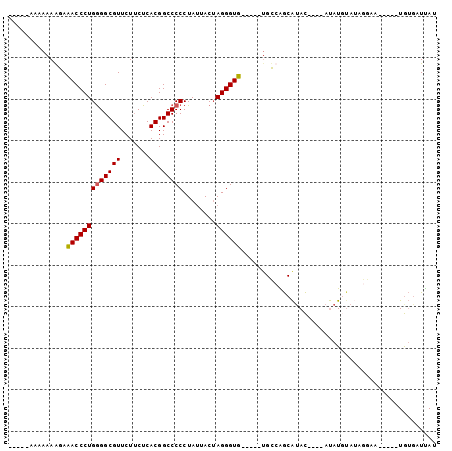
| Location | 5,670,711 – 5,670,801 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.04 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -14.94 |
| Energy contribution | -14.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5670711 90 - 20766785 AUAAUCACA-----UUGAU----AUAU----GUUUGCUGGCUGUUUCAACCCUAGUAAUAGGGGGCCGUGAGAAGAACGCCUCAGGGUUUCUUUUUUUUUUUG ....((((.-----((((.----(((.----(((....)))))).))))(((((....)))))....))))((((((.(((....)))))))))......... ( -22.90) >DroSec_CAF1 8129 84 - 1 AUAAUCACA-----UUCCUAUACAUAU----GUAUGCCGGCA-----CACCCUAGUAAUAGGGGGCCGUGAGAAGAACGCCCCAGGGUAUCCUUUUUU----- ....((((.-----....(((((....----)))))..(((.-----..(((((....))))).)))))))((((.(..((....))..).))))...----- ( -26.80) >DroSim_CAF1 8143 82 - 1 AUAAUCACA-----UUCCUAUACAUAU----GUAUGCUGGCA-----CACCCUAGUAAUAGGGGGCCGUGAGAAGAACGCCCCAGGGUAUCCUUU--U----- ....((((.-----....(((((....----)))))..(((.-----..(((((....))))).)))))))((((.(..((....))..).))))--.----- ( -26.50) >DroYak_CAF1 8217 95 - 1 AUGAUUUUAUUGUAUUUUUUUAAAUGUAUUAAUACGUCUACA-----AACCCUUGUAGUAGGGGGCCGUGAGUAGAACGCCCCAGGGUUUUUUUUUUU---UG ...........(((((..(........)..)))))......(-----((((((........(((((.((.......))))))))))))))........---.. ( -18.60) >consensus AUAAUCACA_____UUCCUAUACAUAU____GUAUGCCGGCA_____AACCCUAGUAAUAGGGGGCCGUGAGAAGAACGCCCCAGGGUAUCCUUUUUU_____ ................................................(((((........(((((.((.......))))))))))))............... (-14.94 = -14.75 + -0.19)

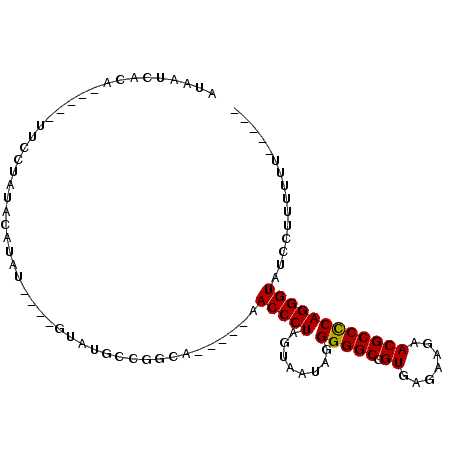
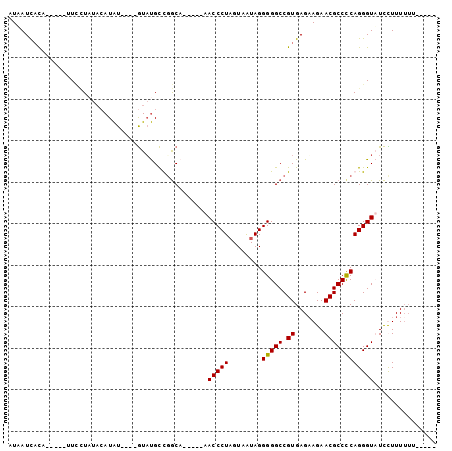
| Location | 5,670,746 – 5,670,836 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.69 |
| Mean single sequence MFE | -17.95 |
| Consensus MFE | -8.27 |
| Energy contribution | -8.78 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5670746 90 - 20766785 AACCAAAA---CAAAAGAAUUAUAUCAUAUAUAUUUCAAUAAUCACA-----UUGAU----AUAU----GUUUGCUGGCUGUUUCAACCCUAGUAAUAGGGGGCCG ........---..............((.((((((.(((((......)-----)))).----))))----)).))..((((.......(((((....))))))))). ( -19.31) >DroSec_CAF1 8159 89 - 1 AACCAAAA---CAAAAGAGUUAUAUCAAAUAUAUUUCAAUAAUCACA-----UUCCUAUACAUAU----GUAUGCCGGCA-----CACCCUAGUAAUAGGGGGCCG ........---.....(((.(((((...))))).)))..........-----....(((((....----))))).((((.-----..(((((....))))).)))) ( -19.60) >DroSim_CAF1 8171 89 - 1 AACCAAAA---CAAAAGAGUUAUAUCAAAUAUAUUUGAAUAAUCACA-----UUCCUAUACAUAU----GUAUGCUGGCA-----CACCCUAGUAAUAGGGGGCCG ........---.....((.((((.(((((....)))))))))))...-----....(((((....----)))))..(((.-----..(((((....))))).))). ( -19.80) >DroYak_CAF1 8249 96 - 1 ACAAA-AAAAAUCAAAA-AUUGUU---UACAAAUUACGAUGAUUUUAUUGUAUUUUUUUAAAUGUAUUAAUACGUCUACA-----AACCCUUGUAGUAGGGGGCCG .....-...........-.((((.---.(((((.(((((((....))))))).))).......(((....)))))..)))-----).((((......))))..... ( -13.10) >consensus AACCAAAA___CAAAAGAAUUAUAUCAAAUAUAUUUCAAUAAUCACA_____UUCCUAUACAUAU____GUAUGCCGGCA_____AACCCUAGUAAUAGGGGGCCG ...........................................................................((((........(((((....))))).)))) ( -8.27 = -8.78 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:12 2006