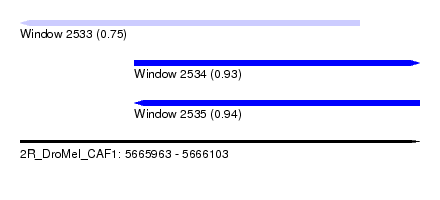
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,665,963 – 5,666,103 |
| Length | 140 |
| Max. P | 0.938567 |

| Location | 5,665,963 – 5,666,082 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -25.38 |
| Energy contribution | -24.94 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5665963 119 - 20766785 UUUC-CUAAAGAUUCCUGAUGGUGUUCACCUGCCUGGCGCUCAGUGUGUUUUCCACCAUCAAGGAGUACGAAGAGGACGCCGUCUACAUUCUGUUCCGCAUGGAGAUCCUGGUGGUUAUC ..((-((...((((((((((((((..((((((.........))).))).....))))))).)))))).)....))))((((((((.(((.(......).))).))))...))))...... ( -34.00) >DroPse_CAF1 3224 118 - 1 --UCUUUCCAGAUUCCUGAUGGUGUUCACAUGCCUCGCCCUGAGCGUGUUUUCCACCAUCAAGGAGUACGAGGACGACGCCGUGUACAUUCUCUUUCGGAUGGAGAUCCUGGUUGUCAUC --.....((((..(((((((((((...((((((.((.....))))))))....))))))).))))(((((.((......)).)))))..(((((.......)))))..))))........ ( -39.00) >DroWil_CAF1 3206 119 - 1 GUUU-UUACAGAUUUCUGAUGGUGUUUACAUGUCUCGCAUUGAGCGUGUUUUCAACCAUCAGGGAAUACGAAGAAGAUGCUGUGUAUAUAUUAUUUAGAAUGGAAAUUUUAGUUGUUAUA ....-.(((((.(..((((((((....(((((.(((.....)))))))).....))))))))..)...(......)...)))))..((((....(((((((....)))))))....)))) ( -30.70) >DroMoj_CAF1 3682 119 - 1 CUC-UUUAUAGAUUUCUGAUGGUGUUUACAUGUCUCGCAUUGAGCGUGUUUUCCACCAUCAAGGAAUAUGAGGAGGAUGCUGUAUAUAUAUUAUUUCGCAUGGAGAUCAUAGUUGUCAUC (((-((((((..((..((((((((...(((((.(((.....))))))))....))))))))..)).)))))))))((((....(((..(((.(((((.....))))).)))..))))))) ( -35.30) >DroAna_CAF1 3423 120 - 1 AUAUAUCUCAGAUUCCUGAUGGUGUUCACCUGCCUGGCUCUGAGCGUAUUCUCCACCAUCAAGGAGUACGAGGAGGACGCCGUCUACAUCCUGUUCCGAAUGGAGAUCCUGGUUGUCAUC .....((((..(((((((((((((....((.....))....(((......)))))))))).))))))..))))..(((((((......(((..........))).....))).))))... ( -32.80) >DroPer_CAF1 3055 118 - 1 --UCCUUUCAGAUUCCUGAUGGUGUUCACAUGCCUCGCCCUGAGCGUGUUUUCCACCAUCAAGGAGUACGAGGACGACGCCGUGUACAUUCUCUUUCGGAUGGAGAUCCUGGUUGUCAUC --(((((....(((((((((((((...((((((.((.....))))))))....))))))).))))))..))))).(((((((.(.....(((((.......))))).).))).))))... ( -41.30) >consensus _UUC_UUACAGAUUCCUGAUGGUGUUCACAUGCCUCGCACUGAGCGUGUUUUCCACCAUCAAGGAGUACGAGGAGGACGCCGUGUACAUUCUAUUUCGAAUGGAGAUCCUGGUUGUCAUC ...........(((((((((((((...(((((.(((.....))))))))....))))))).))))))((.((((.....((((................))))...)))).))....... (-25.38 = -24.94 + -0.44)


| Location | 5,666,003 – 5,666,103 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -20.07 |
| Energy contribution | -19.35 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5666003 100 + 20766785 GCGUCCUCUUCGUACUCCUUGAUGGUGGAAAACACACUGAGCGCCAGGCAGGUGAACACCAUCAGGAAUCUUUAG-GAAAUGAGCAAUCACAAAUUAUUGU--- ((.((((........((((.(((((((.....(((.(((..(....).))))))..)))))))))))......))-)).....))................--- ( -25.64) >DroVir_CAF1 3720 96 + 1 GCAUCCUCCUCGUAUUCUUUGAUGGUGGAAAACACGCUCAAUGCGAGACAUGUGAACACCAUCAGAAAACUGCAAA-AA----ACAAUACAAAAAUAUUAC--- ...........(((((.((((((((((....(((.((((.....))).).)))...)))))))))).)).)))...-..----..................--- ( -21.20) >DroPse_CAF1 3264 90 + 1 GCGUCGUCCUCGUACUCCUUGAUGGUGGAAAACACGCUCAGGGCGAGGCAUGUGAACACCAUCAGGAAUCUGGAAAGA----CAAGGUUACUAC---------- ..(((.(((......((((.(((((((....(((.(((........))).)))...)))))))))))....)))..))----)...........---------- ( -27.70) >DroSec_CAF1 3404 100 + 1 GCGUCCUCUUCGUACUCCUUGAUGGUGGAAAACACACUGAGCGCCAGGCAGGUGAACACCAUCAGGAAUCUUUAG-AAAAUUAAAAAUUACAAUUCAUUGU--- ......(((......((((.(((((((.....(((.(((..(....).))))))..)))))))))))......))-)........................--- ( -22.00) >DroSim_CAF1 3420 99 + 1 GCGUCCUCUUCGUACUCCUUGAUGGUGGAAAACACACUGAGCGCCAGGCAGGUGAACACCAUCAGGAAUCUUUAG-ACAAAAAGAAAUCGAAAAUUAUUG---- ........((((...((((.(((((((.....(((.(((..(....).))))))..))))))))))).(((((..-....)))))...))))........---- ( -25.10) >DroYak_CAF1 3265 104 + 1 GCGUCCUCUUCGUACUCCUUGAUGGUGGAAAACACACUGAGCGCCAGGCAGGUGAACACCAUCAGGAAUCUAAAAAAAAAUGAGAAAUCAAAAGAUUUCGUGAA (((...((((.....((((.(((((((.....(((.(((..(....).))))))..))))))))))).............(((....))).))))...)))... ( -27.30) >consensus GCGUCCUCUUCGUACUCCUUGAUGGUGGAAAACACACUGAGCGCCAGGCAGGUGAACACCAUCAGGAAUCUUUAA_AAAAU_AAAAAUCAAAAAUUAUUGU___ .................((((((((((.....(((.(((.....)))....)))..))))))))))...................................... (-20.07 = -19.35 + -0.72)


| Location | 5,666,003 – 5,666,103 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.32 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5666003 100 - 20766785 ---ACAAUAAUUUGUGAUUGCUCAUUUC-CUAAAGAUUCCUGAUGGUGUUCACCUGCCUGGCGCUCAGUGUGUUUUCCACCAUCAAGGAGUACGAAGAGGACGC ---((((....))))...........((-((...((((((((((((((..((((((.........))).))).....))))))).)))))).)....))))... ( -26.70) >DroVir_CAF1 3720 96 - 1 ---GUAAUAUUUUUGUAUUGU----UU-UUUGCAGUUUUCUGAUGGUGUUCACAUGUCUCGCAUUGAGCGUGUUUUCCACCAUCAAAGAAUACGAGGAGGAUGC ---...........(((((.(----((-((....(((((.((((((((...(((((.(((.....))))))))....)))))))).)))))..))))).))))) ( -26.70) >DroPse_CAF1 3264 90 - 1 ----------GUAGUAACCUUG----UCUUUCCAGAUUCCUGAUGGUGUUCACAUGCCUCGCCCUGAGCGUGUUUUCCACCAUCAAGGAGUACGAGGACGACGC ----------.........(((----(((((....(((((((((((((...((((((.((.....))))))))....))))))).))))))..))))))))... ( -32.70) >DroSec_CAF1 3404 100 - 1 ---ACAAUGAAUUGUAAUUUUUAAUUUU-CUAAAGAUUCCUGAUGGUGUUCACCUGCCUGGCGCUCAGUGUGUUUUCCACCAUCAAGGAGUACGAAGAGGACGC ---((((....)))).........((((-((....(((((((((((((..((((((.........))).))).....))))))).))))))....))))))... ( -25.10) >DroSim_CAF1 3420 99 - 1 ----CAAUAAUUUUCGAUUUCUUUUUGU-CUAAAGAUUCCUGAUGGUGUUCACCUGCCUGGCGCUCAGUGUGUUUUCCACCAUCAAGGAGUACGAAGAGGACGC ----..............((((((((((-......(((((((((((((..((((((.........))).))).....))))))).))))))))))))))))... ( -27.80) >DroYak_CAF1 3265 104 - 1 UUCACGAAAUCUUUUGAUUUCUCAUUUUUUUUUAGAUUCCUGAUGGUGUUCACCUGCCUGGCGCUCAGUGUGUUUUCCACCAUCAAGGAGUACGAAGAGGACGC .....((((((....))))))....((((((((..(((((((((((((..((((((.........))).))).....))))))).))))))..))))))))... ( -31.00) >consensus ___ACAAUAAUUUGUGAUUUCU_AUUUU_CUAAAGAUUCCUGAUGGUGUUCACCUGCCUGGCGCUCAGUGUGUUUUCCACCAUCAAGGAGUACGAAGAGGACGC ...................................(((((((((((((..(((....(((.....))).))).....))))))).))))))............. (-18.70 = -18.32 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:10 2006