
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,653,746 – 5,653,846 |
| Length | 100 |
| Max. P | 0.624587 |

| Location | 5,653,746 – 5,653,846 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -16.21 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5653746 100 - 20766785 UAACUGUUGACGCGCUUCAGCAGUGCGAAGUAGCUGUAGGAGCAGUUGACGAUCUGAAAGGAGAAGAGCAUAACCAGU--A--AUAAAUGAUAUAUAAAGGAAU ..(((((..((..(((((.(((((........))))).))))).))..))..(((....))).............)))--.--..................... ( -23.00) >DroVir_CAF1 25428 103 - 1 UAACUGUUGACACGCUUGAGCAGAGCAAAGUAGCUGUAGGAGCAGUUCACAAUCUGUAAAUUUAAAAUGAUUAUUAGUAUGAUAUG-AUUAAUUAAUAAGCACA .............(((((.(((((.....((((((((....)))))).))..)))))..........(((((((.........)))-)))).....)))))... ( -17.90) >DroSec_CAF1 10644 99 - 1 UAACUGUUGACGCGCUUCAGCAGUGCGAAGUAGCUGUAGGAGCAGUUAACAAUCUGAAAGAAAAAGACCAUAACUAGA--A--AUU-GUGAAAUAUAAAGGACC ....(((((((..(((((.(((((........))))).))))).))))))).................(((((.....--.--.))-))).............. ( -22.10) >DroSim_CAF1 10839 99 - 1 UAACUGUUGACGCGCUUCAGCAGUGCGAAGUAGCUGUAGGAGCAGUUAACAAUCUGAAAGAAAAAGAGGAUAACUAGU--A--AUU-GUGAAAUAUAAAGGACU ....(((((((..(((((.(((((........))))).))))).)))))))((((............)))).......--.--...-................. ( -21.30) >DroEre_CAF1 10443 98 - 1 UAACUGUUGACACGCUUUAGCAGAGCGAAGUAGCUGUAGGAGCAAUUAACAAUCUGAAAGAAAAACAAUAUAAAUAGU--U--AUU-GUGAUAU-UAAAGGACU ....((((((...((((..((((..........))))..))))..)))))).(((.........((((((........--)--)))-)).....-....))).. ( -17.77) >DroYak_CAF1 10492 99 - 1 UAACUGUUGACGCGUUUCAGCAGUGCGAAGUAGCUGUAAGAACAGUUAACAAUCUGAAAGAAAAAGAACAUAACUAGU--U--AUU-GAGCUAUAUAAAGGACU ..((((((((......)))))))).....((((((.(((.((((((((....(((.........)))...))))).))--)--.))-))))))).......... ( -21.10) >consensus UAACUGUUGACGCGCUUCAGCAGUGCGAAGUAGCUGUAGGAGCAGUUAACAAUCUGAAAGAAAAAGAACAUAACUAGU__A__AUU_GUGAAAUAUAAAGGACU ....(((((((..(((((.(((((........))))).))))).)))))))..................................................... (-16.21 = -16.60 + 0.39)

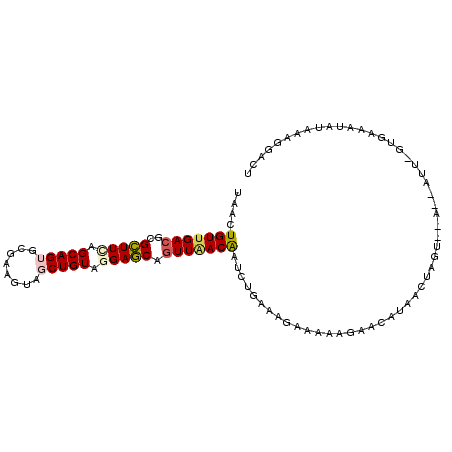
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:03 2006