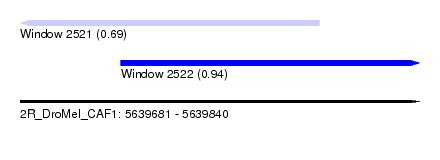
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,639,681 – 5,639,840 |
| Length | 159 |
| Max. P | 0.935462 |

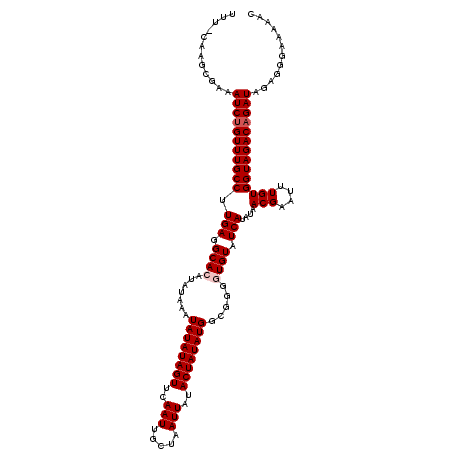
| Location | 5,639,681 – 5,639,800 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5639681 119 - 20766785 UACACCCCGCCAUAUAGUAUAAUUAGCAAUUGAACUAUAUAUUUAUAUGUGCCUCAAGGCAAACGGAUUUCGCUUG-AAAAAUGUAGUCACUCAAUUGUAGGGAGCUUUAAAGGCGGCUG ......(((((....(((....(..(((((((((((((((.........((((....))))......((((....)-))).)))))))...))))))))..)..))).....)))))... ( -29.00) >DroSec_CAF1 6374 119 - 1 UACACCCCGCCAUAUAGUAUAAUUAGCAAUUGAACUAUAUAUUUAUAUGUGCCUCAAGGCAAACAGAUUUCGCUUG-AAAAAUGUAGUCACUCAAUUGUAGGGAGCUUUAAAGGCGGCUG ......(((((....(((....(..(((((((((((((((.........((((....))))......((((....)-))).)))))))...))))))))..)..))).....)))))... ( -29.00) >DroSim_CAF1 6444 119 - 1 UACACCCCGCCAUAUAGUAUAAUUAGCAAUUGAACUAUAUAUUUAUAUGUGCCUCAAGGCAAACAGAUUUCGCUCG-AAAAAUGUAGUCACUCAAUUGUAGGGAGCUUUAAAGGCGGCUG ......(((((....(((....(..(((((((((((((((.........((((....))))......((((....)-))).)))))))...))))))))..)..))).....)))))... ( -29.90) >DroEre_CAF1 6799 118 - 1 UACACCCCGCCAUAUAGUAUAAUUAGCAAUUGAACUAUAUAUUUAUAUUUGCCUCAAGGCAAACAGAUUUUGCUUG-AAAAAUGUAAUCACCCA-CUGAAAGGAGCUUUAAAGGCGGCUG ......((((((((((((.((((.....)))).)))))))..((((((((...(((((.((((.....))))))))-).)))))))).......-((....)).........)))))... ( -26.90) >DroYak_CAF1 6396 120 - 1 UACACCCCGCCAUAUAGUAUAAUUAGCAAUUGAACUAUAUAUUUAUAUUUGCCUCAAGGCAAACAGAUUUUGCUUGGAAAAAUGUAAUCACCCAAUUGUAAGGAGCUUGGGAGGCGACAG .......(((((((((((.((((.....)))).)))))))..((((((((...((.(((((((.....))))))).)).))))))))...(((((..((.....))))))).)))).... ( -29.00) >consensus UACACCCCGCCAUAUAGUAUAAUUAGCAAUUGAACUAUAUAUUUAUAUGUGCCUCAAGGCAAACAGAUUUCGCUUG_AAAAAUGUAGUCACUCAAUUGUAGGGAGCUUUAAAGGCGGCUG ......((((((((((((.((((.....)))).))))))).........((((....))))..........((((....((.((........)).)).....))))......)))))... (-22.02 = -22.26 + 0.24)

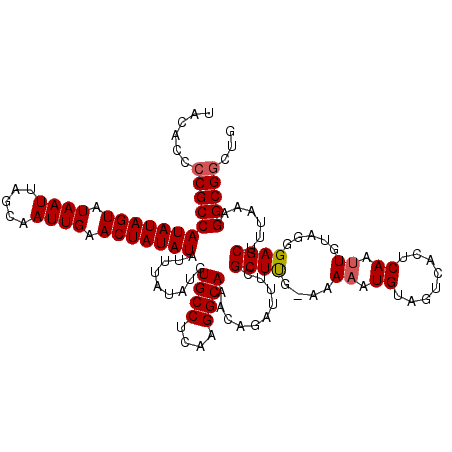
| Location | 5,639,721 – 5,639,840 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5639721 119 + 20766785 UUU-CAAGCGAAAUCCGUUUGCCUUGAGGCACAUAUAAAUAUAUAGUUCAAUUGCUAAUUAUACUAUAUGGCGGGGUGUAUCAUAUAACGAAUUUGUGGUAGACAGAUAGUGGGAAAAAC (((-(..((....(((((.((((....))))((((((..((((((((......))))..)))).)))))))))))((.(((((((.........))))))).)).....))..))))... ( -28.90) >DroSec_CAF1 6414 119 + 1 UUU-CAAGCGAAAUCUGUUUGCCUUGAGGCACAUAUAAAUAUAUAGUUCAAUUGCUAAUUAUACUAUAUGGCGGGGUGUAUCAUAUAACGAAUUUGUGGUAGACAGAUAGAGGGAAAAAC (((-(...(...(((((((((((.(((.((((.......((((((((..(((.....)))..)))))))).....)))).)))....(((....)))))))))))))).)...))))... ( -29.00) >DroSim_CAF1 6484 119 + 1 UUU-CGAGCGAAAUCUGUUUGCCUUGAGGCACAUAUAAAUAUAUAGUUCAAUUGCUAAUUAUACUAUAUGGCGGGGUGUAUCAUAUAACGAAUUUGUGGUAGACAGAUAGAGGGAAAAAC (((-(...(...(((((((((((.(((.((((.......((((((((..(((.....)))..)))))))).....)))).)))....(((....)))))))))))))).)...))))... ( -27.90) >DroEre_CAF1 6838 119 + 1 UUU-CAAGCAAAAUCUGUUUGCCUUGAGGCAAAUAUAAAUAUAUAGUUCAAUUGCUAAUUAUACUAUAUGGCGGGGUGUAUCAUAUAACGAAAUUGUGGUAGACAGAUAGAGGGAAAAAC (((-(..((......((((((((....))))))))....((((((((..(((.....)))..))))))))))...((.(((((((.........))))))).)).........))))... ( -26.50) >DroYak_CAF1 6436 120 + 1 UUUCCAAGCAAAAUCUGUUUGCCUUGAGGCAAAUAUAAAUAUAUAGUUCAAUUGCUAAUUAUACUAUAUGGCGGGGUGUAUCAUAUAACGAAAUUGUGGUAGACAGAUAGAGGGAAAAAC (((((..((......((((((((....))))))))....((((((((..(((.....)))..))))))))))...((.(((((((.........))))))).))........)))))... ( -29.80) >consensus UUU_CAAGCGAAAUCUGUUUGCCUUGAGGCACAUAUAAAUAUAUAGUUCAAUUGCUAAUUAUACUAUAUGGCGGGGUGUAUCAUAUAACGAAUUUGUGGUAGACAGAUAGAGGGAAAAAC ............(((((((((((.(((.((((.......((((((((..(((.....)))..)))))))).....)))).)))....(((....))))))))))))))............ (-23.98 = -24.58 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:56 2006