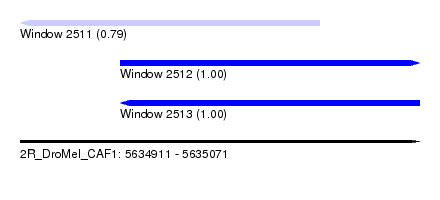
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,634,911 – 5,635,071 |
| Length | 160 |
| Max. P | 0.998830 |

| Location | 5,634,911 – 5,635,031 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -22.87 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5634911 120 - 20766785 CUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAACGUAAUAUGAAGUGAAUGGAGCUACAGGUCUGAAAAUUAGCACGG ...........(((((((((((.(((((.......))))).)))))))))))...((((((((...(((((.....))))).((.(((.......))).))))).......))))).... ( -26.41) >DroSec_CAF1 1619 120 - 1 CUUAAAAAUACUCUUUAUGGGCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAGCGUAAUAUGAAGUGAAUGGAGCUACAGGUCUGAAAAUUAGCACGG ...........(((((((((.(.(((((.......))))).).)))))))))...((((((((...(((((.....))))).((.(((.......))).))))).......))))).... ( -21.81) >DroSim_CAF1 1617 120 - 1 CUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAGCGUAAUAUGAAGUGAAUGGAGCUACAGGUCUGAAAAUUAGCACGG ...........(((((((((((.(((((.......))))).)))))))))))...((((((((...(((((.....))))).((.(((.......))).))))).......))))).... ( -26.41) >DroEre_CAF1 1953 120 - 1 CUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAAAUGUCGGCGGUCAUACAGAAAAGUUAAACCUAAAUUAUAAAGCGUAAUAUGAAGUGAAUGGAGCUACAGGUCUCGAAAUUAGCACGG ...........(((.(((((((.(((((.......))))).))))))).)))...(((((.......(((((........)))))........((((.....).)))....))))).... ( -19.70) >DroYak_CAF1 1595 119 - 1 CUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCUAAAUUA-AAAGCGUAAUAUAAAGUGAAUGGAGCCACAGGUCUGGAAAUUAGCACGG (((((......(((((((((((.(((((.......))))).)))))))))))....))))).........-...............(((.......(((......))).......))).. ( -27.84) >consensus CUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAGCGUAAUAUGAAGUGAAUGGAGCUACAGGUCUGAAAAUUAGCACGG ...........(((((((((((.(((((.......))))).)))))))))))...((((((((....(((((........))))).(((........))).))).......))))).... (-22.87 = -22.83 + -0.04)

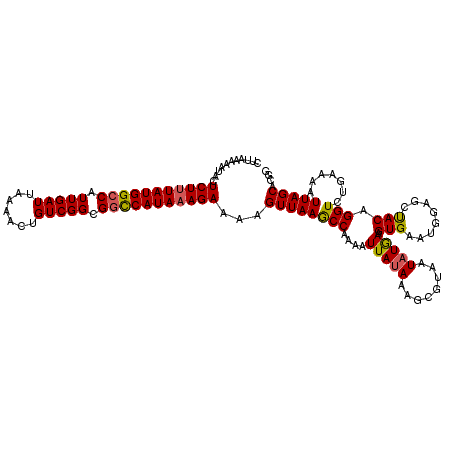
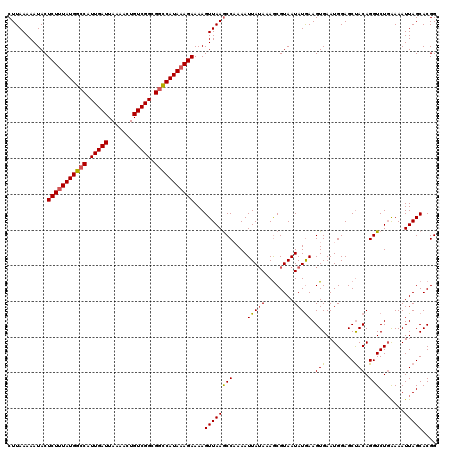
| Location | 5,634,951 – 5,635,071 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -23.66 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5634951 120 + 20766785 UUACGUUUUAUAAUUUUGGCUUAACUUUUCUUUAUGGCCGCCGACAGUUUUAAUCAAUGGCCAUAAAGAGUAUUUUUAAGCCAAAUGAUGCUAAUUUUUUGGAAUUGAUAGCGCUAAUUU ...((((...((((((((((((((...(((((((((((((..((.........))..))))))))))))).....)))))))))......((((....)))))))))..))))....... ( -31.70) >DroSec_CAF1 1659 97 + 1 UUACGCUUUAUAAUUUUGGCUUAACUUUUCUUUAUGGCCGCCGACAGUUUUAAUCAAUGCCCAUAAAGAGUAUUUUUAAGCCAAAUGAUGCUAA-----------------------UUU ....((.((((....(((((((((...((((((((((.((..((.........))..)).)))))))))).....))))))))))))).))...-----------------------... ( -23.20) >DroSim_CAF1 1657 120 + 1 UUACGCUUUAUAAUUUUGGCUUAACUUUUCUUUAUGGCCGCCGACAGUUUUAAUCAAUGGCCAUAAAGAGUAUUUUUAAGCCAAAUGAUGCUAAUUUUUUGGAAUUGAUAGCGCUAAUUU ...((((...((((((((((((((...(((((((((((((..((.........))..))))))))))))).....)))))))))......((((....)))))))))..))))....... ( -34.10) >DroEre_CAF1 1993 120 + 1 UUACGCUUUAUAAUUUAGGUUUAACUUUUCUGUAUGACCGCCGACAUUUUUAAUCAAUGGCCAUAAAGAGUAUUUUUAAGCCAAAUGAUGUUAAUUUAUGGGAAUUAAAAGCGCUAAUUU ...((((((.((((((.((((..((......))..)))).(((.........((((.((((..((((((...)))))).))))..)))).........)))))))))))))))....... ( -22.97) >DroYak_CAF1 1635 118 + 1 UUACGCUUU-UAAUUUAGGCUUAACUUUUCUUUAUGGCCGCCGACAGUUUUAAUCAAUGGCCAUAAAGAGUAUUUUUAAGCCAAAUGAUGCUAAUUUGUA-UAAUUAAGAGCGCUAAUUU ...((((((-(((((..(((((((...(((((((((((((..((.........))..))))))))))))).....))))))).....((((......)))-))))))))))))....... ( -39.40) >consensus UUACGCUUUAUAAUUUUGGCUUAACUUUUCUUUAUGGCCGCCGACAGUUUUAAUCAAUGGCCAUAAAGAGUAUUUUUAAGCCAAAUGAUGCUAAUUUUUUGGAAUUAAUAGCGCUAAUUU ....((.(((....((((((((((...(((((((((((((..((.........))..))))))))))))).....))))))))))))).))............................. (-23.66 = -24.38 + 0.72)

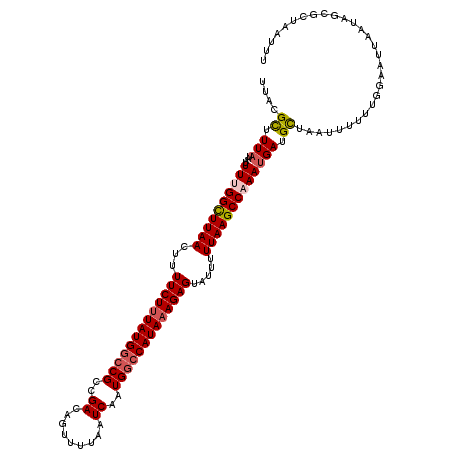
| Location | 5,634,951 – 5,635,071 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -27.52 |
| Energy contribution | -28.36 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -5.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5634951 120 - 20766785 AAAUUAGCGCUAUCAAUUCCAAAAAAUUAGCAUCAUUUGGCUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAACGUAA ......((((((...............))))....((((((((((......(((((((((((.(((((.......))))).)))))))))))....))))))))))..........)).. ( -36.26) >DroSec_CAF1 1659 97 - 1 AAA-----------------------UUAGCAUCAUUUGGCUUAAAAAUACUCUUUAUGGGCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAGCGUAA ...-----------------------.........((((((((((......(((((((((.(.(((((.......))))).).)))))))))....)))))))))).............. ( -28.30) >DroSim_CAF1 1657 120 - 1 AAAUUAGCGCUAUCAAUUCCAAAAAAUUAGCAUCAUUUGGCUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAGCGUAA ......((((((...............))))....((((((((((......(((((((((((.(((((.......))))).)))))))))))....))))))))))........)).... ( -38.46) >DroEre_CAF1 1993 120 - 1 AAAUUAGCGCUUUUAAUUCCCAUAAAUUAACAUCAUUUGGCUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAAAUGUCGGCGGUCAUACAGAAAAGUUAAACCUAAAUUAUAAAGCGUAA ......((((((((((((........(((((...((((.......))))..(((.(((((((.(((((.......))))).))))))).)))...))))).....))))).))))))).. ( -24.32) >DroYak_CAF1 1635 118 - 1 AAAUUAGCGCUCUUAAUUA-UACAAAUUAGCAUCAUUUGGCUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCUAAAUUA-AAAGCGUAA ......(((((.((((((.-...((((.......))))(((((((......(((((((((((.(((((.......))))).)))))))))))....)))))))..)))))-).))))).. ( -37.40) >consensus AAAUUAGCGCUAUCAAUUCCAAAAAAUUAGCAUCAUUUGGCUUAAAAAUACUCUUUAUGGCCAUUGAUUAAAACUGUCGGCGGCCAUAAAGAAAAGUUAAGCCAAAAUUAUAAAGCGUAA ...................................((((((((((......(((((((((((.(((((.......))))).)))))))))))....)))))))))).............. (-27.52 = -28.36 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:46 2006