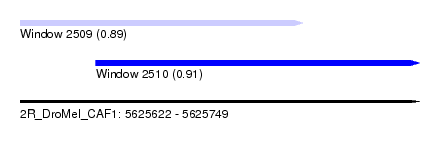
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,625,622 – 5,625,749 |
| Length | 127 |
| Max. P | 0.912361 |

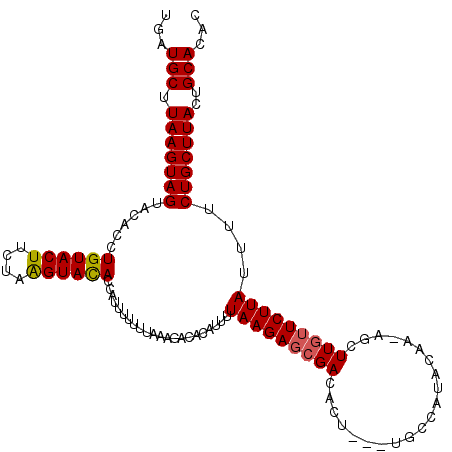
| Location | 5,625,622 – 5,625,712 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5625622 90 + 20766785 --UACUUCAGUGCAGUUGUCGUUGCUUGAUGCUUAAGUAGUACACCUGU----------ACACAAUUUUUUGAAAGACACAUUUUAAGAGCGACACU---UGCCA --.......(.((((.(((((((((((((...)))))))((((....))----------))......((((((((......)))))))))))))).)---)))). ( -22.30) >DroSec_CAF1 818 103 + 1 --UACUUCAGUGCAGUUGUCGUUGCUUGAUGCUUAAGUAGUACACCUGUACUUCUAAGUACACCAUUUUUUUAAAGACACAUUUUAAGAGCGACACAAAGAGCCA --.......(.((..((((.(((((((..((((((.(.(((((....))))).)))))))..........((((((.....)))))))))))))))))...))). ( -22.60) >DroSim_CAF1 764 100 + 1 --UACUUCAGUGCAGUUGUCGUUGCUUGAUGCUUAAGUAGUACACAUGUACUUCUAAGUACACCAUUUUUUUAAAGACACAUUUUAAGAGCGACACU---UGCCA --.......(.((((.(((((((.(((((((((((.(.(((((....))))).))))))).....((((....))))......)))))))))))).)---)))). ( -24.70) >DroEre_CAF1 857 100 + 1 --UACUUCAGUGCUGUUGUCGUUGCUUAAUGCUUAAGUAGUACACUUGUACUUUUAAGUAUACCAUUUUUUAAGACACAAUUUUUAAGAGCGACACU---UGCCA --.......(.((...(((((((.....(((((((((.(((((....)))))))))))))).......(((((((......))))))))))))))..---.))). ( -24.30) >DroYak_CAF1 847 102 + 1 CAUACUUCAGUGCAGUUGUCGUUGCUUAAUGCUUAAGUAGUACACUUGUACUUUUAGGUACACCAUUUUUUUAAAGACACUAUUUAAGAGCGACACU---UGCCA .........(.((((.(((((((.(((((((((((((.(((((....))))))))))))).....((((....))))......)))))))))))).)---)))). ( -26.10) >consensus __UACUUCAGUGCAGUUGUCGUUGCUUGAUGCUUAAGUAGUACACCUGUACUUCUAAGUACACCAUUUUUUUAAAGACACAUUUUAAGAGCGACACU___UGCCA .........(.(((..(((((((.(((((((((.....))))....((((((....)))))).....................)))))))))))).....)))). (-17.41 = -17.44 + 0.03)

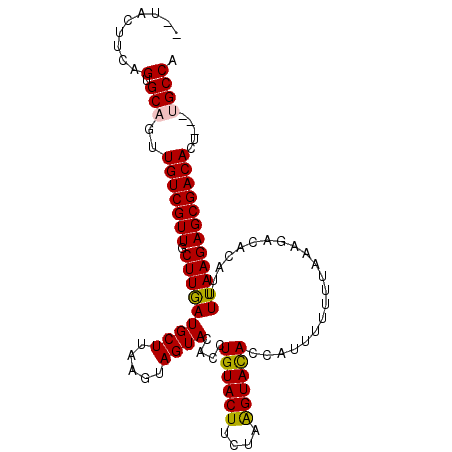
| Location | 5,625,646 – 5,625,749 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5625646 103 + 20766785 UGAUGCUUAAGUAGUACACCUGU----------ACACAAUUUUUUGAAAGACACAUUUUAAGAGCGACACU---UGCCAUACAAACGCUUGUUCUUAUUUUCUGCUUACUGCACAC ((.(((.((((((((((....))----------)).......................((((((((((..(---((.....)))..).))))))))).....))))))..))))). ( -18.80) >DroSec_CAF1 842 114 + 1 UGAUGCUUAAGUAGUACACCUGUACUUCUAAGUACACCAUUUUUUUAAAGACACAUUUUAAGAGCGACACAAAGAGCCAUA-AA-AGCUUGAUCUUAUUUUCUGCUUACUGCACAC ((.(((.(((((((......((((((....))))))...........((((((..(((((.(.((..........))).))-))-)...)).)))).....)))))))..))))). ( -19.90) >DroSim_CAF1 788 112 + 1 UGAUGCUUAAGUAGUACACAUGUACUUCUAAGUACACCAUUUUUUUAAAGACACAUUUUAAGAGCGACACU---UGCCAUACAA-AGCUUGUUCUUAUUUUCUGCUUACUGCACAC ((.(((.(((((((......((((((....))))))......................(((((((((..((---(........)-)).)))))))))....)))))))..))))). ( -23.40) >DroEre_CAF1 881 112 + 1 UAAUGCUUAAGUAGUACACUUGUACUUUUAAGUAUACCAUUUUUUAAGACACAAUUUUUAAGAGCGACACU---UGCCAUACAA-AGCUUAUUCUUAUUUUCUGCUUACUGCACAC ..(((((((((.(((((....)))))))))))))).........((((.((.((....((((((..(..((---(........)-)).)..))))))..)).))))))........ ( -18.20) >DroYak_CAF1 873 112 + 1 UAAUGCUUAAGUAGUACACUUGUACUUUUAGGUACACCAUUUUUUUAAAGACACUAUUUAAGAGCGACACU---UGCCAUACAA-AGCUUGUUCUUAUUUUCUGCUUACUGCACAC ...(((.(((((((......((((((....))))))......................(((((((((..((---(........)-)).)))))))))....)))))))..)))... ( -23.40) >consensus UGAUGCUUAAGUAGUACACCUGUACUUCUAAGUACACCAUUUUUUUAAAGACACAUUUUAAGAGCGACACU___UGCCAUACAA_AGCUUGUUCUUAUUUUCUGCUUACUGCACAC ...(((.(((((((......((((((....))))))......................(((((((((.....................)))))))))....)))))))..)))... (-17.01 = -17.28 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:43 2006